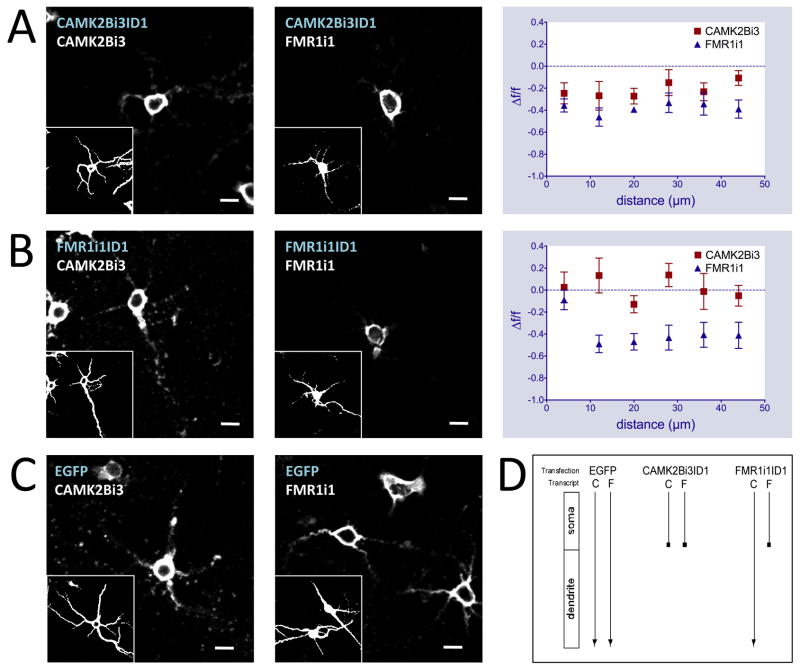
Figure 3.
Intronic ID element sequences disrupt dendritic localization patterns of endogenous mRNA. In situ hybridizations with antisense biotinylated intron riboprobes on primary hippocampal neurons transfected with CAMK2Bi3ID1-EGFP (A), FMR1i1ID1-EGFP (B), pEGFP-N1 (C) constructs (GABRG3i5ID2-EGFP and GRIK1i1ID4 results found in Figure S4). Blue text indicates transfected DNA construct, white text indicates in situ probe sequence. Graphs at right represent in situ signal F/F against distance from soma for ID-EGFP constructs versus pEGFP-N1 using CAMK2Bi3 (left) and FMR1i1 (right) riboprobes. Insets represent MAP2 immunostaining. Scale bars = 20μm. (D) Schematic of ID cross competition results. Transfection labels indicate transfected DNA constructs, transcript labels indicate endogenous intron-retaining transcripts: C is CAMK2Bi3, F is FMR1i1. Arrows indicate endogenous intron-retaining transcript targeting from soma to dendrites.