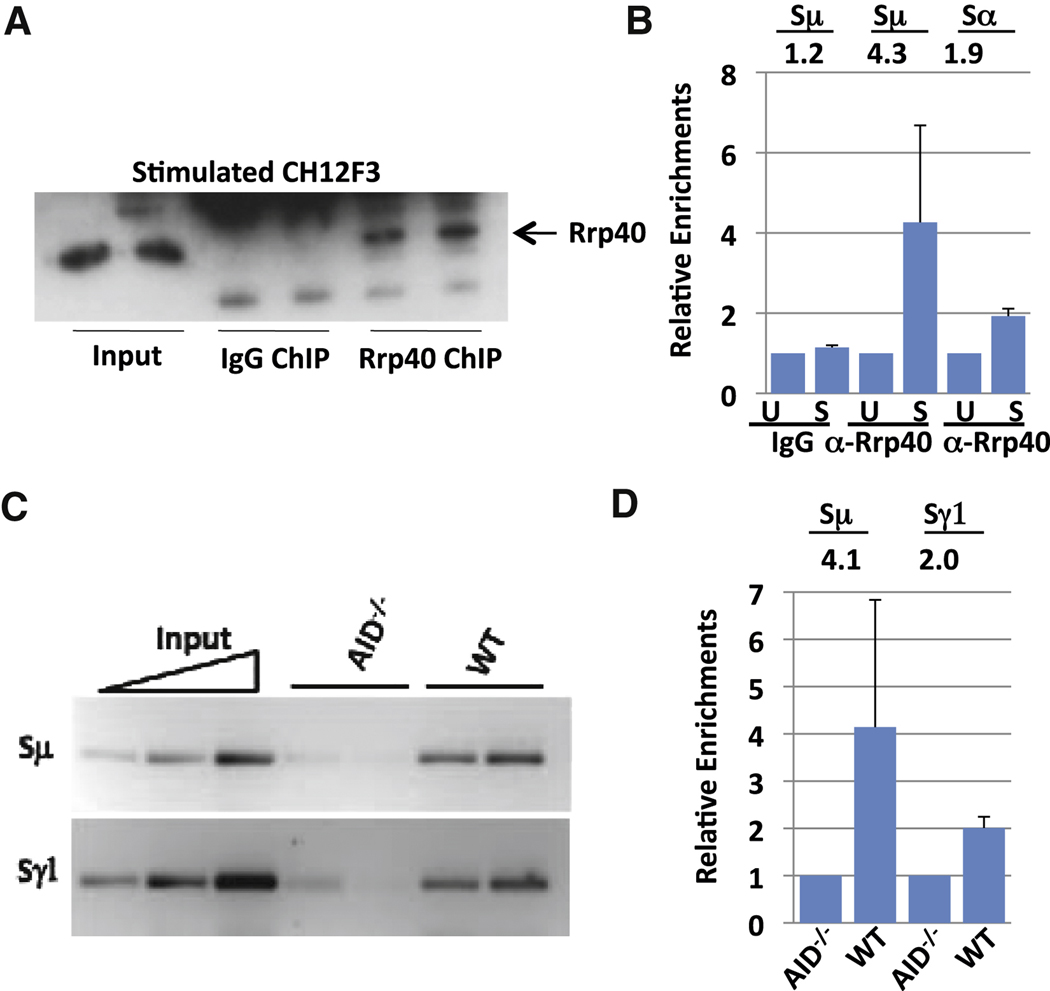
Figure 4. RNA exosome subunit Rrp40 is recruited to S regions.
(A) Ch12F3 cells were either stimulated with TGFβ, IL4 and CD40 or kept un-stimulated for 48 hours. Subsequently, Rrp40 was immunoprecipitated from cell extracts under chromatin immunoprecipitation (ChIP) conditions and immunoprecipitates analyzed for Rrp40 by Western blotting with anti-Rrp40. (B) The "ChIPed" Rrp40-DNA complex from Ch12F3 cells was processed to isolate bound DNA. ChIPed DNA was tested for Sµ and Sα sequences via q-PCR. The average and standard deviation from the mean for three separate ChIP experiments is shown (see also Fig. S4). Numbers indicate average fold changes comparing stimulated and un-stimulated samples. Un-stimulated samples were arbitrarily normalized as 1 (Supp. Methods). (C) Rrp40 ChIPs were performed on extracts from primary splenic B cells stimulated for 2 days with anti-CD40 plus IL4. Sµ and Sγ1 were tested via semi-quantitative PCR; results are shown for two independent ChIP samples for each genotype. A 5-fold serial dilution of inputs is shown with the highest input concentration corresponding to 1/20 of total input. (D) ChIPed DNA from activated splenic B cells was tested for Sµ and Sγ1 via q-PCR. Numbers indicate average fold changes comparing WT and AID−/− samples. AID−/− samples were arbitrarily normalized as 1 (Supp. Methods). Values represent the average and standard deviation from the mean for three experiments. See Figure S4 for more details.