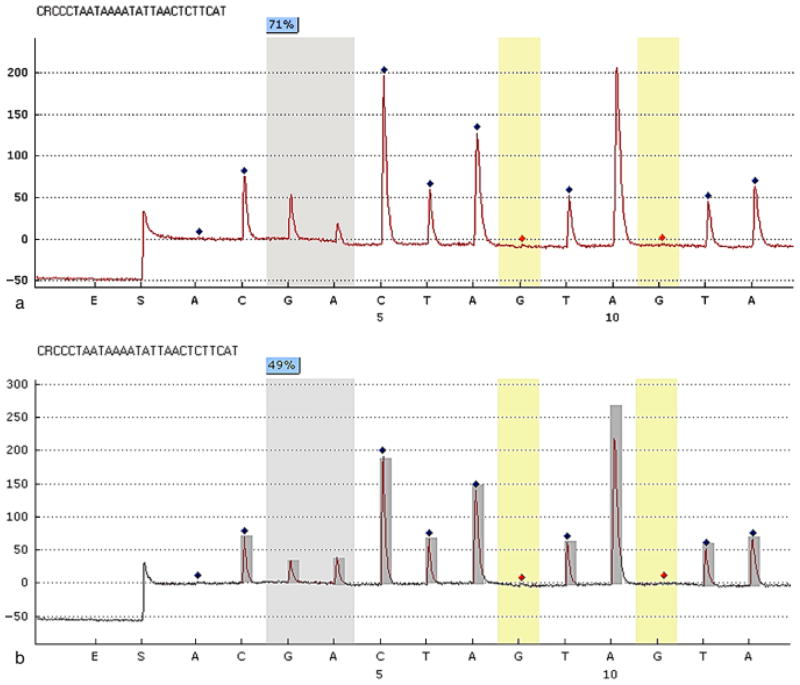
Fig. 2.
Representative programs of pyrosequencing. The percentage on each CpG site is the methylation percentage of mC/(mC+C) on that site. mC, methylated cytosine; C, unmethylated cytosine. The sequence shown on the top of each panel is the assayed sequence. The x-axes are the dispensation nucleotides to the sequencing reaction based on the assayed sequences. The y-axes show light emission obtained as relative light units. The methylation percentage of one specific CpG site (− 295 bp) from a healthy gingival biopsy sample (a), which shows the value of methylation percentage of 71, and a chronic periodontitis sample (b), which shows a value of methylation percentage of 49 are illustrated. This percentage is calculated from the reference peak heights, marked as dark diamonds, which are non-CpG nucleotide sequences. The red diamond marks indicate “built-in” bisulphite controls in pyrosequencing.