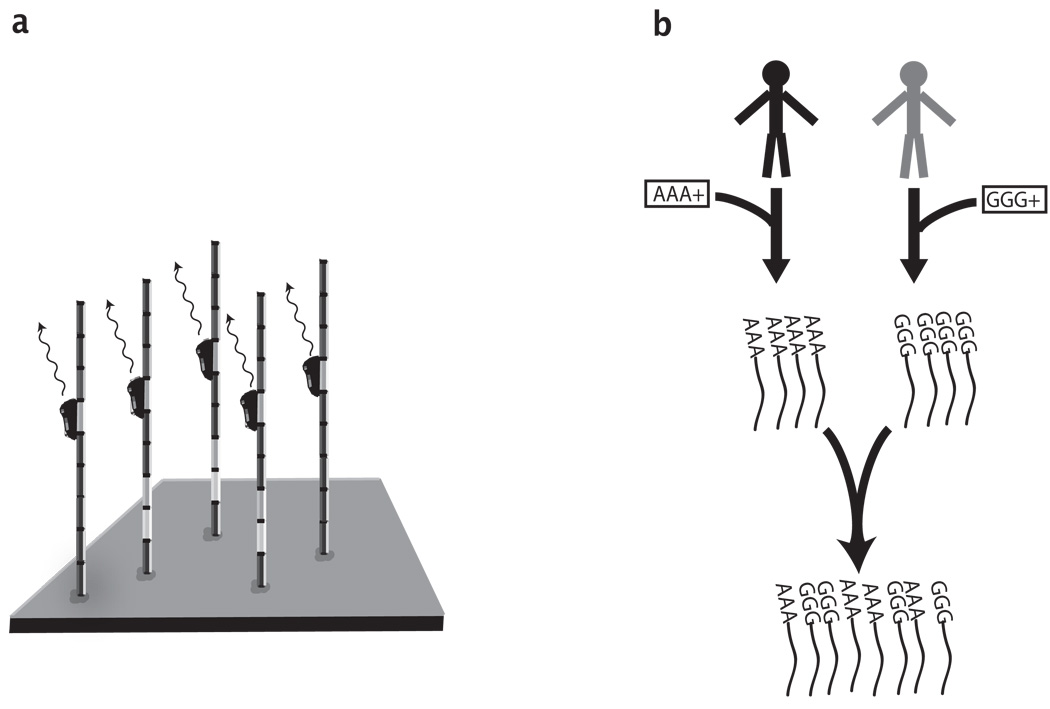
Fig. 1.
Common techniques in next generation sequencing (a) Schematic Overview of High throughput DNA Sequencing. The DNA fragments (vertical rods) are immobilized onto a surface and occupy distinct spatial locations. The sequencing reagents (black ovals) generate optical signals according to the DNA composition of each fragment. A series of signals from the same spatial location conveys the sequence of a single DNA fragment. (b) DNA Barcoding. DNA barcoding starts with synthesizing short DNA sequences. In the example, there are two barcodes: ’AAA’ and ’GGG’. The barcodes are then concatenated in a simple chemical reaction to the DNA fragments of each sample (black lines). When the barcoded samples are mixed and sequenced, the barcodes retain the origin of the sequences. In this example, every sequence read will start with a short tag of 3 nucleotides, either ’AAA’ or ’GGG’.