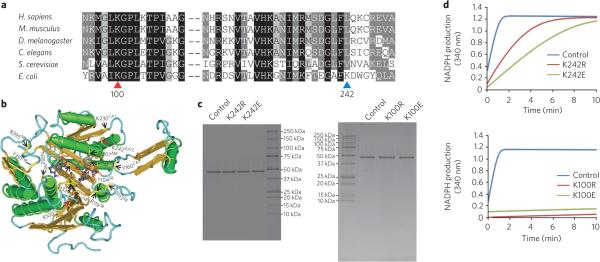
Figure 6. Sequence alignment and mutagenesis analysis of isocitrate dehydrogenase.
(a) ClustalW (2.0.12) alignment of isocitrate dehydrogenase homologs from H. sapiens (GenInfo Identifier (GI): 5031777), M. musculus (GI: 148693874), D. melanogaster (GI: 24643268), C. elegans (GI: 71986051), S. cerevisiae (GI: 6324709) and E. coli (GI: 170080787). Conserved sites are shaded with gray and black. Conserved and nonconserved succinyllysine residues are indicated by red and blue triangles, respectively. The positions are labeled corresponding to the E. coli sequence. (b) localization of the succinyllysines and functional sites in isocitrate dehydrogenase. The three-dimensional structure was obtained from the Molecular Modeling Database (MMDB) (MMDB ID 49631) and viewed by Cn3D (v4.1). Succinylated and known functionally important sites (Uniprot ID P08200) are indicated by red and yellow arrows, respectively. The superscripts SB, NB, MB, CAT and SUCC on the labeled residues refer to: substrate binding, NADP binding, metal binding, catalytic sites and succinylation, respectively. (c) Purity of the wild-type and mutated proteins shown by SDS-PAGE gel. (d) Enzymatic activities of wild-type (control), K242R, K242E (top), K100E and K100R (bottom) isocitrate dehydrogenase mutants.