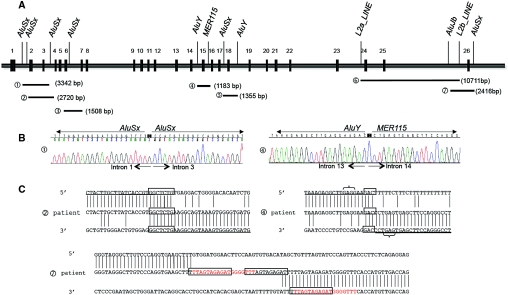
Figure 4.
Mapping and characterization of breakpoints of SLC12A3 heterozygous deletions. (A) Schematic representation of the genomic organization of the 26 exons of the SLC12A3 gene and the location of breakpoints of 7 deletions: (A1) E2_E3del: 3342 bp deletion (c.282 + 667_c.506–205del). (A2) E2_E3del: 2720 bp deletion (c.283–273_c.506–213del). (A3) E4_E6del: 1508 bp deletion (c.506–315_852 + 185del). (A4) E14del: 1183 bp deletion (c.1169 + 773_c.1825 + 247del). (A5) E18del: 1355 bp deletion (c.2178 + 269_c.2285 + 685del). (A6) E24_E25del: 10711bp deletion (c.2748–324_c.2952–505). (A7) E26del: 2416bp deletion (c.2952–1593_*677delins25). The LCRs on or near the breakpoints are indicated. (B) Sequence analysis showing breakpoints of deletions 1 and 4. Breakpoints of deletion 1 are inside Alu repeats, suggesting a nonallelic homologous recombination. Breakpoints of deletion 4 are inside nonhomologous LCRs. (C) Sequence alignments at the breakpoints of two SLC12A3 heterozygous deletions probably originated from nonhomologous end-joining and one complex rearrangement. For deletions 2 and 4, boxes indicate the nucleotide microhomology. Brackets depict short motifs that might have facilitated the rearrangements. For deletion 7, boxes indicate the repetitive motifs; the read sequence is the 18-bp repetition that is present in inserted sequence and in the 3′UTR breakpoint.