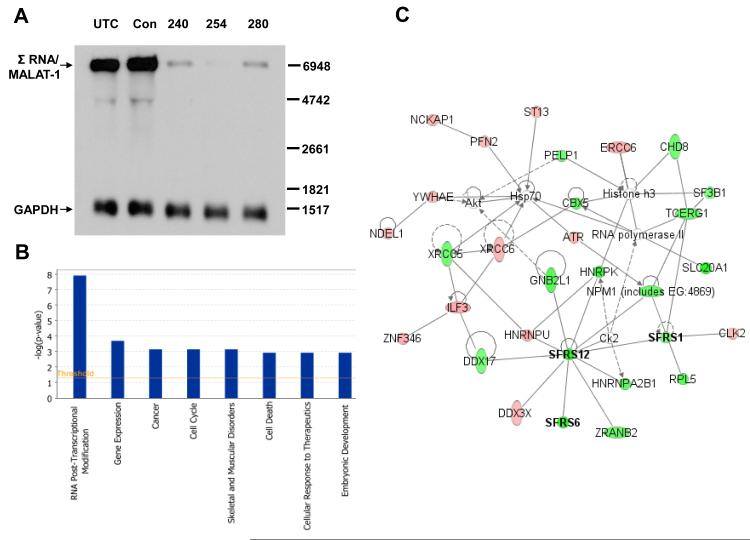
Fig. 1.
Evidence that Σ RNA/MALAT-1 regulates RNA post-transcriptional modification globally in HeLa cells. (A) Target reduction of Σ RNA/MALAT-1 by ASOs. RNAs from HeLa cell transfectants were used for Northern analyses. Position of Σ RNA/MALAT-1 is indicated. GAPDH, loading controls. UTC, untreated control; Con, scrambled ASO; 240, 254, and 280, Σ RNA/MALAT-1 specific ASOs. (B) IPA analyses of genes whose alternative splicing activity were significantly changed (P<0.001) by Σ RNA/MALAT-1 reduction in HeLa cells. (C) Components of the top class of differentially expressed genes and their network. Bold, transcripts of SR proteins. Red and green, up- and down- regulated transcripts, respectively, in the ASO254-transfected HeLa cells with significantly changed alternative splicing activity (P<0.001) due to Σ RNA/MALAT-1 reduction. Uncolored, genes in the network that were not identified in the exon array analysis.