Abstract
Summary: GeneReporter is a web tool that reports functional information and relevant literature on a protein-coding sequence of interest. Its purpose is to support both manual genome annotation and document retrieval. PubMed references corresponding to a sequence are detected by the extraction of query words from UniProt entries of homologous sequences. Data on protein families, domains, potential cofactors, structure, function, cellular localization, metabolic contribution and corresponding DNA binding sites complement the information on a given gene product of interest.
Availability and implementation: GeneReporter is available at http://www.genereporter.tu-bs.de. The web site integrates databases and analysis tools as SOAP-based web services from the EBI (European Bioinformatics Institute) and NCBI (National Center for Biotechnology Information).
Contact: i.retter@tu-bs.de; ida.retter@helmholtz-hzi.de
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
In face of next-generation sequencing and high-throughput analyses, the link between obtained data and existing knowledge is crucial. Automatic annotation pipelines provide useful evidence of potential functions for genes and proteins, but in a last essential step, the scientist must manually evaluate the available information. Usually, the necessary evidence is derived from scientific publications, databases and in silico predictions. Thus, tools that provide a combination of all of these relevant data for a gene or protein of interest are of high practical impact. In this context, GeneReporter offers a customizable workflow for the integrated application of protein sequence analysis and document retrieval.
A large number of diverse text-mining tools exist that provide different strategies and interfaces to satisfy the extensive data-mining demands in biomedical sciences (Krallinger et al., 2010). GeneReporter identifies citations related to a gene or protein sequence of interest. The UniProt annotations of homologous sequences are used to derive keywords such as gene names, synonyms and species. These keywords provide the query terms for a subsequent literature search in PubMed (Sayers et al., 2010). In this way, GeneReporter extends and replaces MineBlast (Dieterich et al., 2005), a similar tool which is discontinued. In comparison with other tools that connect literature to sequence information, like quickLit (Gilchrist et al., 2008) and Metis (Mitchell et al., 2005), GeneReporter is characterized by highly customizable query options, the integration of InterPro and the direct access to the original EBI and NCBI databases.
2 REQUESTING LITERATURE AND SEQUENCE ANALYSIS
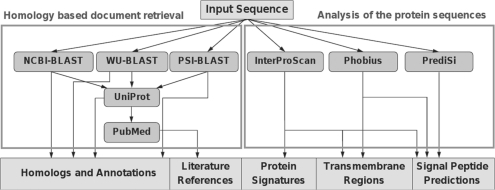
The user can enter up to 10 nt or protein sequences to submit a query on the GeneReporter web site. Two different types of analyses are provided: (i) homology-based document retrieval searches information on homologous sequences from the UniProt Knowledgebase (UniProt Consortium, 2010) and citations from PubMed. (ii) Analysis of the protein sequences requests protein annotations from InterPro (Hunter et al., 2009), Phobius (Käll et al., 2007) and PrediSi (Hiller et al., 2004). The complete workflow is depicted in Figure 1. An example for an application is given as Supplementary Material.
Fig. 1.
Workflow of the GeneReporter analysis process. Arrows indicate data transfer and processing. Input and output is depicted as rectangles, web services are depicted as rounded rectangles.
Using homology-based document retrieval, the first step is a BLAST search in UniProtKB, where the user can select the desired algorithm. NCBI-BLAST (Altschul et al., 1997) and WU-BLAST (Lopez et al., 2003) result in a different ranking of homology matches, and therefore yield different query word extractions from the respective UniProtKB entries. PSI-BLAST (Altschul et al., 1997) is the most sensitive algorithm and beneficial for sequences that fail to result in significant hits with the other algorithms. Either Swiss-Prot or the complete UniProtKB can be chosen as BLAST target database. The UniProtKB entries of the resulting BLAST hits are parsed for gene names, synonyms and species names, which are used as query terms for the subsequent PubMed request. This literature search can be further specified, e.g. by additional query terms and years of publication. The option ‘organism-specific search’ adds the respective species name to the PubMed search string. Query and result options and the construction of the PubMed queries are described in detail in the Supplementary Material.
For further analysis of the protein sequence, GeneReporter submits a query to InterProScan that matches the sequence against InterPro. This database comprises predictive signatures that assign protein families, various domains and functional sites for a protein of interest. The input sequence can also be analysed by Phobius and PrediSi, which search for putative transmembrane regions and signal peptides.
To assure long-term up-to-date datasets and analysis tools, GeneReporter utilizes standardized web services from the EBI (Goujon et al., 2010), the NCBI (Sayers et al., 2010) and our institute. The processing time of a query strongly depends on these services. The web service providers bind the access of their services to certain rules in order to avoid overload and abuse of their resources. To match these rules, a local queuing system monitors and limits the number of simultaneous queries. Details on cut-offs and limits are provided as Supplementary Material.
3 RESULTS
The results are summarized on an overview page. For each query sequence, this page provides a link to a detailed view of the obtained data for the requested services. The result overview page can be bookmarked and results can be retrieved from this URL for at least 24 h. For further analysis, results can be downloaded as Excel or tab-delimited text files.
The detailed view provides one result tab for each requested service. The BLAST result tab shows homologous protein sequences. It is complemented with annotations from the UniProt database, e.g. organism name and GO terms, in order to facilitate their evaluation. The PubMed result tab shows gene-related citations ordered by the respective PubMed queries. Query words that were matched within title and abstract are marked in bold. For each query word combination, the link ‘This query in PubMed’ performs the corresponding query on the PubMed web site. This allows the manual modification and specification of the automatically generated queries with all the sophisticated features of the PubMed search interface. Furthermore, GeneReporter provides citations from UniProt entries of the BLAST hit sequences. In general, these references comprise the key papers on the respective gene or protein. Figure 2 shows the PubMed tab of an example search for a hypothetical protein from Pseudomonas aeruginosa C3719.
Fig. 2.
Screenshot of the homology-based document retrieval result. Query sequence in this example: UniProt AcNo A3KZR4.
The output from InterPro, Phobius and PrediSi requests is given in additional tabs. The InterProScan and Phobius output includes graphical visualizations of signature matches and transmembrane regions within the proteins of interest.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank the EBI and NCBI for providing web service access to their tools and databases. We would also like to thank Max Schobert for intensive testing and discussions and Patrick Jäkel for the layout of the GeneReporter logo.
Funding: Grants of the German Federal Ministry of Education and Research (BMBF) for the European transnational research initiative on ‘Systems Biology of Microorganisms’, SysMO (Psysmo) (grant number 0313980D); the Volkswagen Foundation (I/81448).
Conflict of Interest: none declared.
REFERENCES
- Altschul S.F., et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterich G., et al. MineBlast: a literature presentation service supporting protein annotation by data mining of BLAST results. Bioinformatics. 2005;21:3450–3451. doi: 10.1093/bioinformatics/bti528. [DOI] [PubMed] [Google Scholar]
- Gilchrist M.J., et al. Evading the annotation bottleneck: using sequence similarity to search non-sequence gene data. BMC Bioinformatics. 2008;9:442. doi: 10.1186/1471-2105-9-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goujon M., et al. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 2010;38:W695–W699. doi: 10.1093/nar/gkq313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller K., et al. PrediSi: prediction of signal peptides and their cleavage positions. Nucleic Acids Res. 2004;32:W375–W379. doi: 10.1093/nar/gkh378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter S., et al. InterPro: the integrative protein signature database. Nucleic Acids Res. 2009;37:D211–D215. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez R., et al. WU-Blast2 server at the European Bioinformatics Institute. Nucleic Acids Res. 2003;31:3795–3798. doi: 10.1093/nar/gkg573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Käll L., et al. Advantages of combined transmembrane topology and signal peptide prediction–the Phobius web server. Nucleic Acids Res. 2007;35:W429–W432. doi: 10.1093/nar/gkm256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krallinger M., et al. Analysis of biological processes and diseases using text mining approaches. Methods Mol. Biol. 2010;593:341–382. doi: 10.1007/978-1-60327-194-3_16. [DOI] [PubMed] [Google Scholar]
- Mitchell A.L., et al. METIS: multiple extraction techniques for informative sentences. Bioinformatics. 2005;21:4196–4197. doi: 10.1093/bioinformatics/bti675. [DOI] [PubMed] [Google Scholar]
- Sayers E.W., et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2010;38:D5–D16. doi: 10.1093/nar/gkp967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt Consortium. The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res. 2010;38:D142–D148. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.