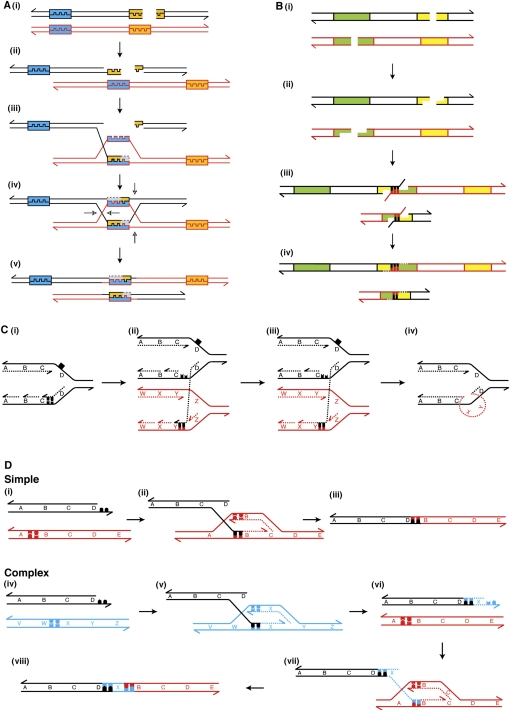
Figure 4.
Schematic representations of postulated mechanisms for the formation of genomic rearrangements. In all panels, 3′ ends are represented by half arrows. (A) Non-allelic homologous recombination. (i) A DSB occurs. (ii) The DNA ends are resected by exonucleases leaving 3′ single-stranded DNA (ssDNA) overhangs. (iii) One of the 3′ ssDNA ends invades a paralogous sequence (blue and orange boxes) and is extended by DNA synthesis. (iv) Strand invasion and extension of the second DNA end occurs, resulting in the formation of a double Holliday junction. (v) The Holliday junctions are resolved in the opposite sense producing a reciprocal duplication and deletion. (B) Non-homologous DNA end joining. (i) Two DSBs occur in sister chromatids or homologous chromosomes. (ii) The Artemis–DNA-dependent protein kinase catalytic subunit (DNA-PKcs) complex resects the DNA ends. (iii) Incorrect DNA ends are brought together by the Ku protein, and may anneal through microhomology. The Artemis–DNA–PKcs complex cleaves the flaps. (iv) DNA polymerase λ or μ (POLL or POLM) adds nucleotides to fill the gaps, and the NHEJ1–XRCC4–DNA ligase IV (LIG4) complex ligates the DNA ends, producing a reciprocal deletion and duplication. (C) Fork stalling template switching. (i) A replication fork stalls after encountering a DNA lesion (black square) on the template strand. (ii) The lagging strand from the stalled replication fork anneals to the template strand of another replication fork using microhomology. (iii) The invading strand is either extended by DNA synthesis or ligated to the 5′ end of the preceding Okazaki fragment. (iv) The FoSTeS model predicts that the lagging strand returns to the original template, allowing normal DNA replication to resume. Prior to this occurring, the lagging strand may invade multiple different replication forks (not shown). (D) Microhomology-mediated break-induced replication. (i–iii) Formation of a simple rearrangement by MMBIR. (i) An unpaired DSB end undergoes 5′ resection. (ii) The 3′ overhang uses microhomology to anneal to a non-paralogous section of DNA. (iii) No subsequent template switching events occur. (iv–viii) Formation of a complex rearrangement by MMBIR. (iv,v) Strand invasion occurs as in the creation of a simple rearrangement. (vi) The extended end dissociates as the low-processivity replication fork collapses. (vii) The 3′ end anneals to a second template. For tandem duplications this is upstream of the original break. (viii) Further template switching events may occur (not shown) until ultimately a fully processive replication fork is formed that goes to completion or collides with a replication fork moving in the opposite direction.