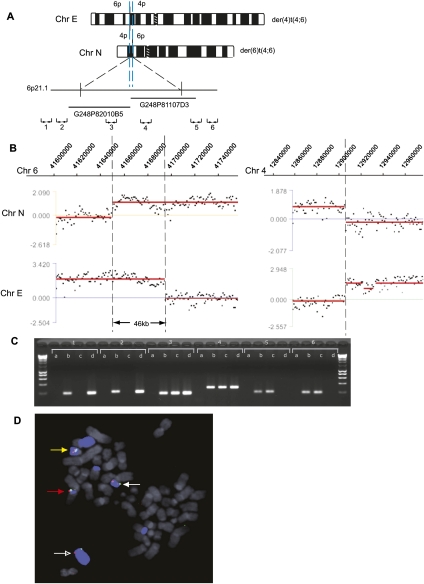
Figure 2.
HCC1806 t(4;6) duplication at a breakpoint. (A) Schematic representation of the products (Chr E and Chr N) of the reciprocal translocation between chromosomes 4 and 6 in HCC1806. The duplicated region of Chr 6 is shown between blue dashed lines, and the approximate location of fosmids used in FISH mapping is shown. The approximate location of chromosome 6 PCR primer pairs (1–6) is also shown. (B) Hybridization of Chr N and Chr E to a custom NimbleGen oligonucleotide array covering specified regions on chromosome 6 (left) and chromosome 4 (right). Breakpoints are indicated with a broken line. (C) Breakpoint mapping by PCR on flow-sorted chromosomes. PCR results with primer pairs 1–6 are shown. Lanes are labeled a (negative water control), b (HCC1806 genomic DNA), c (Chr N), and d (Chr E). (D) Breakpoint mapping by FISH using fosmids G248P81107D3 (shown in red) and G248P82010B5 (shown in green). Chromosome 6 is shown in blue. The der(4)t(1;6;4) is indicated with a red arrow (Chr B), the der(4)t(4;6) with a white arrow (Chr E), the der(6)t(4;6) with an open white arrow (Chr N), and the del (6) with a yellow arrow (Chr j). Other pieces of Chr 6 are the der(10)t(6;10) (Chr V), the der(14)t(6;14) (Chr Z), and the der(6)t(1;6) (Chr F) (Howarth et al. 2008).