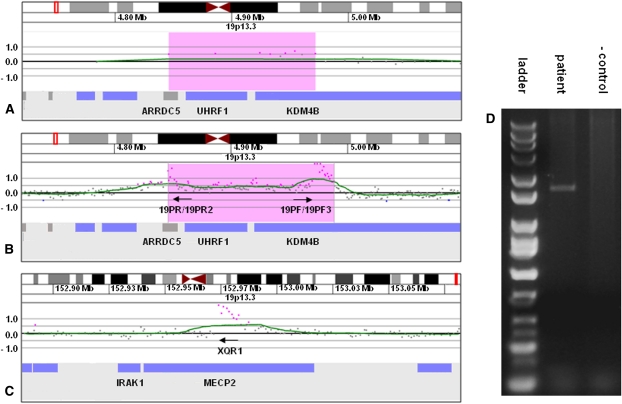
Figure 3.
Characterization of der(X)ins(X;19)(q28;p13.3p13.3) by oligonucleotide array CGH, linear amplification, and PCR. (A) 135k-feature oligonucleotide microarray results showing a single-copy gain of 9 probes, ∼126 kb in size (chr19:4,845,920–4,971,768 based on the UCSC 2006 hg 18 assembly), from 19p13.3 in proband 6. Probes are ordered along the x-axis with the most distal 19p13.3 probes to the left and the most proximal 19p13.3 probes to the right. Values along the y-axis represent log2 ratios of patient:control signal intensities. (B) 2.1M feature oligonucleotide microarray analysis showing the same duplication as in A after linear amplification with primers 19PF, 19PF3, 19PR, and 19PR2. Successful amplification is evidenced by the elevated log ratios of probes at the proximal and distal edges of the duplicated region. (C) 2.1M feature oligonucleotide microarray analysis showing linear amplification from B extending across the insertion junction into intron 2 of MECP2. (D) Gel electrophoresis of PCR amplicon produced with primers 19PF and XQR1 confirming the insertion site detected by linear amplification.