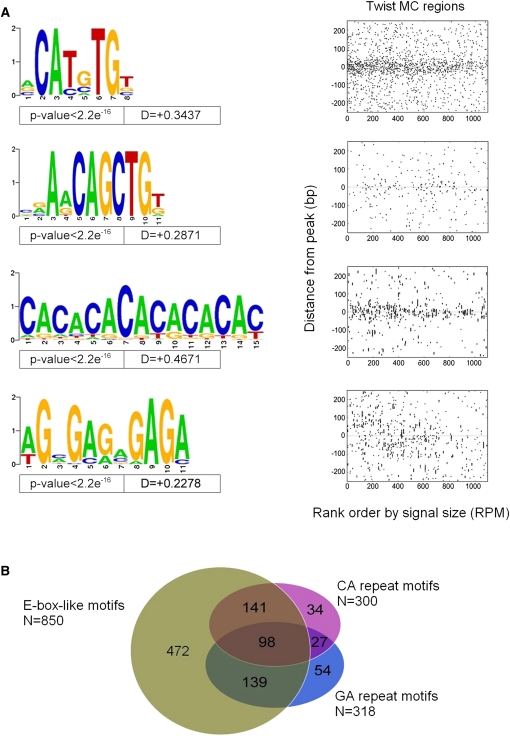
Figure 5.
Motifs associated with Twist in vivo occupancy identified using MEME. MEME was run on the narrow 50 bp region surrounding each of the 1099 MC ChIP-seq peaks to identify all motifs that are enriched near the point of Twist occupancy. These motifs were mapped back to determine their spatial distribution relative to Twist peaks, and some motifs showing a non-uniform distribution near Twist peaks were selected. (A) Variations on CAYRTG and CAGCTG were returned, together specifying CABVTG (top two Weblogos). Note that a leading A residue or a lagging T residue is also suggested, which appears preferred by other non-Twist family DNA-binding bHLH factors (K Fisher-Aylor, S Kuntz, and A Kirilusha, unpubl. obs.; Grove et al. 2009). In addition, two simple repetitive sequences (CA and GA) are also significantly enriched at Twist-occupied sites (bottom two Weblogos). (B) Venn diagram illustrating the relationship between sets of peaks defined as having at least one occurrence of (i) either of the two E-box-like motifs; (ii) the CA-repeat-like sequence; or (iii) the GA-repeat-like sequence.