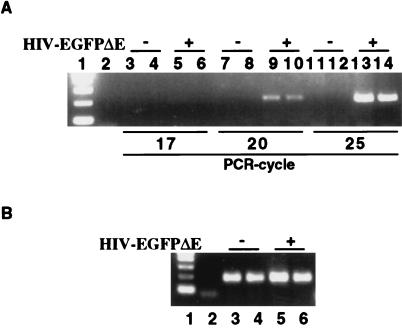
Figure 4.
PCR analysis of HIV-1 DNA in MEFs infected with HIV-EGFPΔE. HIV-1 and cellular GAPDH from infected MEFs were detected by PCR. (A) MEFs were infected with HIV-EGFPΔE for 2 h, after which time DNA was isolated. To detect HIV-1 sequence quantitatively without overamplification, the HIV-1 sequence was amplified for 17, 20, and 25 PCR cycles. Lane 1 represented the 1 kb DNA ladder marker. Lane 2 represented the water control with no DNA at 22 PCR cycles. Lanes 3, 4, 7, 8, 11, and 12 represented MEFs infected with HIV-EGFPΔE for 0 min, where cells were infected with HIV-EGFPΔE but immediately washed three times with 1 × PBS. Lanes 5, 6, 9, 10, 13, and 14 represented cells infected with HIV-EGFPΔE for 2 h. Lanes 3 ≈ 6 represented 17 PCR cycles, 7 ≈ 10, 20 PCR cycles, and 11 ≈ 14, 25 PCR cycles. The odd-numbered lanes represented PARP-1+/+ MEFs, whereas even-numbered lanes represented PARP-1−/− MEFs, except lanes 1 and 2. In quantitative analysis of PCR from 17 to 25 cycles, equal amounts of HIV-1 DNA were detected only in HIV-EGFP-infected PARP-1+/+ and PARP-1−/− MEFs. There was no HIV-1 DNA in the control PCR. (B) The same sample was also amplified for 20 PCR cycles to detect GAPDH, normalizing for total cellular DNA. Lane 1 represented 1-kb DNA ladder and lane 2, water control, lanes 3 and 5, PARP-1+/+ MEFs, and lanes 4 and 6, PARP-1−/− MEFs. Lanes 3 and 4 represented MEFs infected with HIV-EGFPΔE for 0 min and lanes 5 and 6, for 2 h. At 20 PCR cycles, GAPDH DNA was equally amplified, indicating equal amounts of cellular DNA in PCR samples. Amplified PCR products were analyzed by an ethidium-bromide-stained 2% agarose gel.