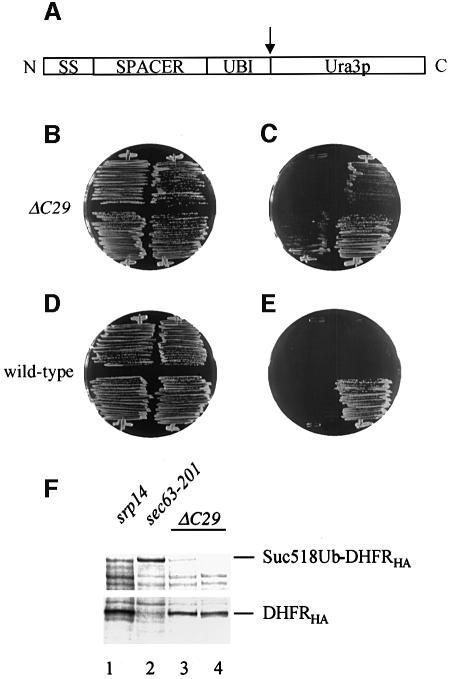
Fig. 8. Translocation defects of srp14-ΔC29 cells in the UTA assay. (A) Organization of UTA constructs. The protein is depicted N- to C-terminus; signal sequence (SS), spacer (SPACER), ubiquitin (UBI) and Ura3p. The cleavage site for cytosolic ubiquitin-dependent proteases is indicated (arrow). (B and C) srp14-ΔC29 cells streaked on to a –trp (B; selecting for the plasmids) and a –ura (C; selecting for cytosolic Ura3p) plate were incubated for 2 days at 30°C. They contained empty vector (top left), Suc2277 (top right), Dap2300 (bottom left) or the control substrate Suc223, which has a 23 amino acid spacer (bottom right). (D and E) As for (B) and (C) but with wild-type cells. As seen previously (Johnsson and Varshavsky, 1994), Suc223 gave a Ura+ phenotype in wild-type cells. (F) Anti-HA immunoprecipitations from extracts of pulse-labelled cells expressing Suc2518UbDHFRHA (lanes 1–3) or Suc223UbDHFRHA control, which only yields the DHFRHA fragment (lane 4) (Johnsson and Varshavski, 1994). Labelling of cells and analysis are as in Figure 7.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.