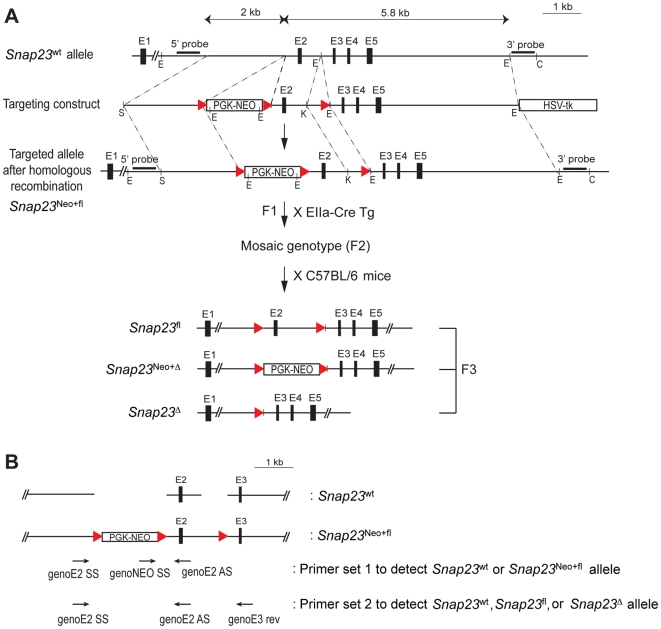
Figure 1. Gene targeting strategy for the generation of Snap23 fl/wt and Snap Δ23/wt mice.
(A)Schematic representation of the genomic structure of wild-type (wt) and targeted alleles of Snap23. Homologous recombination with the targeting construct inserts a neomycin-resistance gene (PKG-NEO) and exon 2 (E2) flanked by three loxP sites represented as triangles. HSK-tk was used for negative selection. Exon numbers with relative position, 5′- and 3′- flanking Southern blotting, and a partial restriction map are indicated. Restriction sites: C, ClaI; E, EcoRI; K, KpnI; S, SpeI. Female Snap23 Neo+fl/wt heterozygous mouse harboring the targeted allele (F1) was bred with male EIIa-Cre transgenic mice. Offspring harboring mosaic alleles (F2) through partial and/or total excision of loxP-flanked sequences were generated. EIIa-Cre+ F2 mosaic male mice were further bred with C57BL/6 female mice to separate the Snap23 fl/wt, Snap23 Neo+fl/wt, and Snap23 Δ/wt alleles as shown (F3). (B) Location of oligonucleotide primers used to discriminate mice harboring different Snap23 alleles following Cre-mediated excision. Genotyping for the Snap23 wt and Snap23 Neo+fl alleles was performed by PCR Primer set 1 and yielded fragments of 266 bp and 400 bp from the Snap23 wt and Snap23 Neo+fl alleles, respectively. The Neo- Snap23 fl and Snap23 Δ alleles were identified by PCR using PCR Primer set 2, which yields fragments of 266 bp, 400 bp, and 492 bp from the Neo- Snap23 wt, Snap23 fl, and Snap23 Δ alleles, respectively (also see Table 1).