Abstract
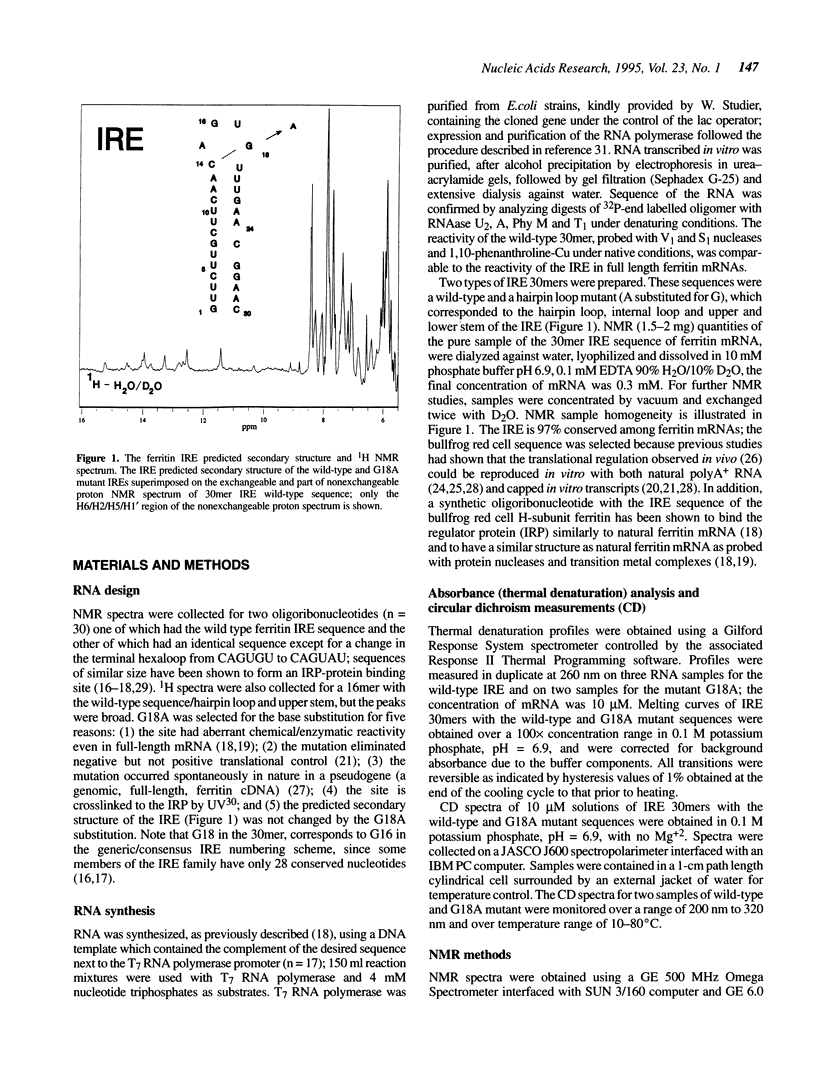
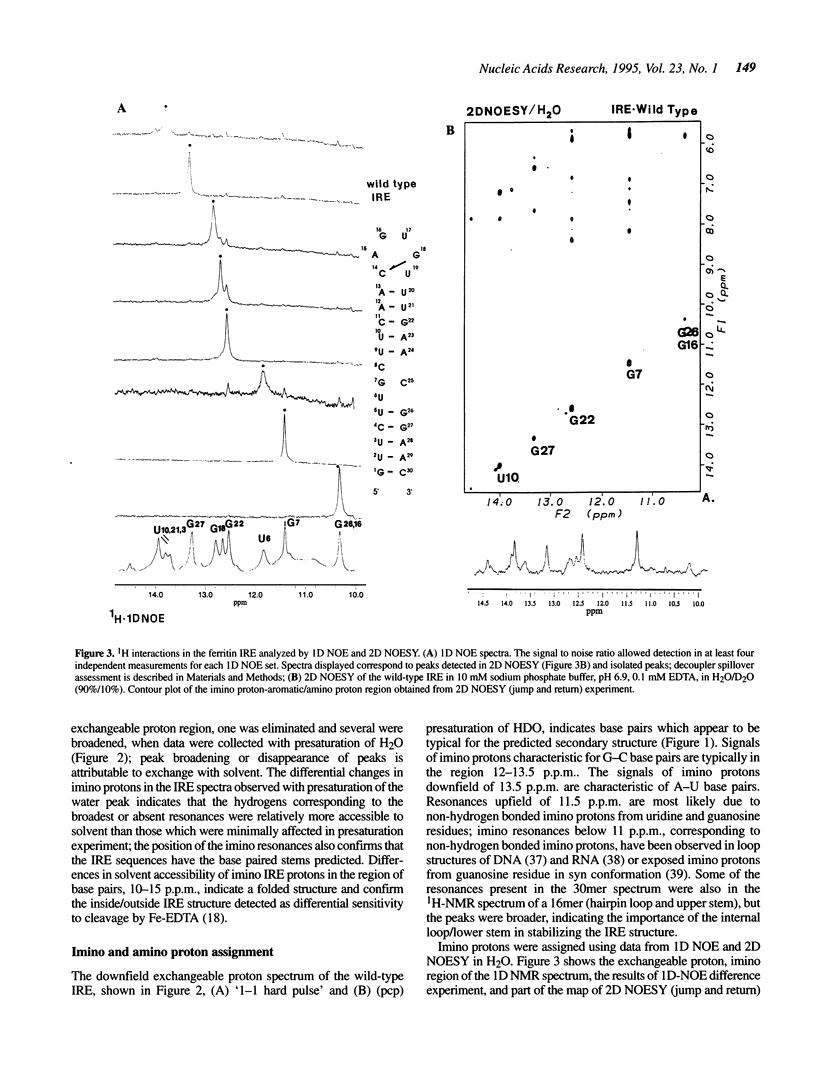
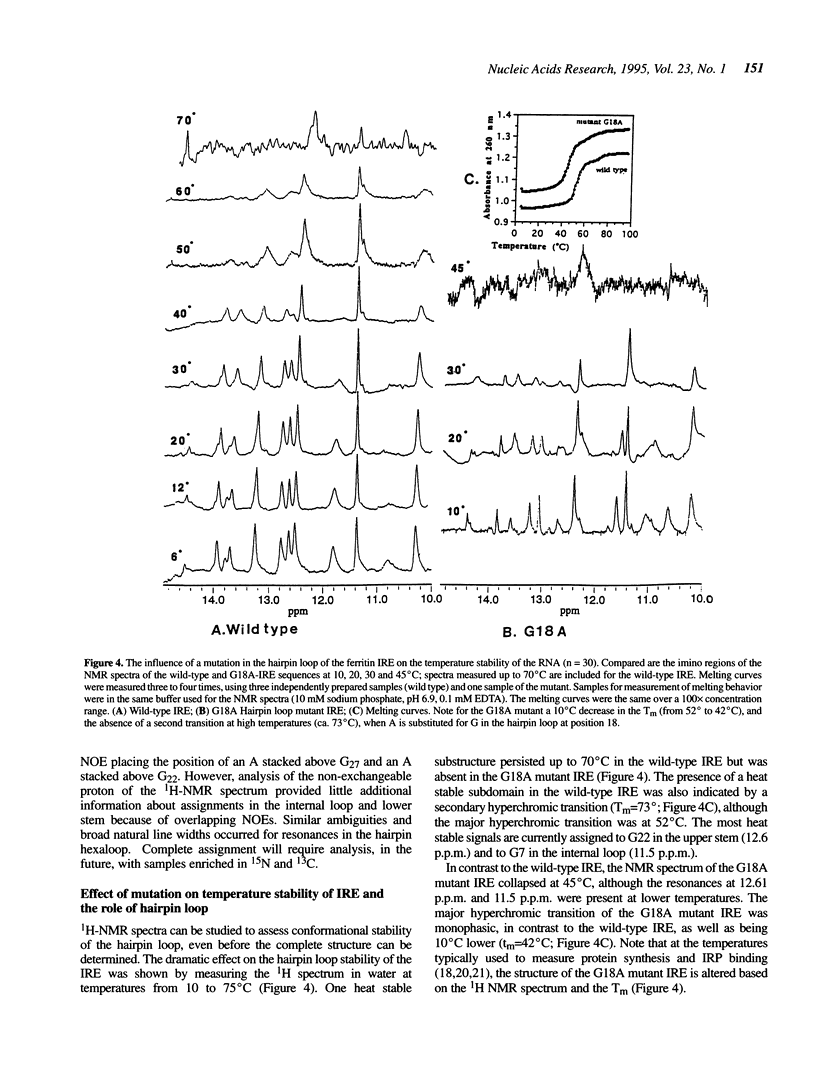
Noncoding sequences regulate the function of mRNA and DNA. In animal mRNAs, iron responsive elements (IREs) regulate the synthesis of proteins for iron storage, uptake and red cell heme formation. Folding of the IRE was indicated previously by reactivity with chemical and enzymatic probes. 1H- and 31P-NMR spectra now confirm the IRE folding; an atypical 31P-spectrum, differential accessibility of imino protons to solvents, multiple long-range NOEs and heat stable subdomains were observed. Biphasic hyperchromic transitions occurred (52 and 73 degrees C). A G-C base pair occurs in the hairpin loop (HL) (based on dimethylsulfate, RNAse T1 previously used, and changes in NMR imino proton resonances typical of G-C base pairs after G/A substitution). Mutation of the hairpin loop also decreased temperature stability and changed the 31P-NMR spectrum; regulation and protein (IRP) binding were previously shown to change. Alteration of IRE structure shown by NMR spectroscopy, occurred at temperatures used in studies of IRE function, explaining loss of IRP binding. The effect of the HL mutation on the IRE emphasizes the importance of HL structure in other mRNAs, viral RNAs (e.g. HIV-TAR), and ribozymes.
Full text
PDF



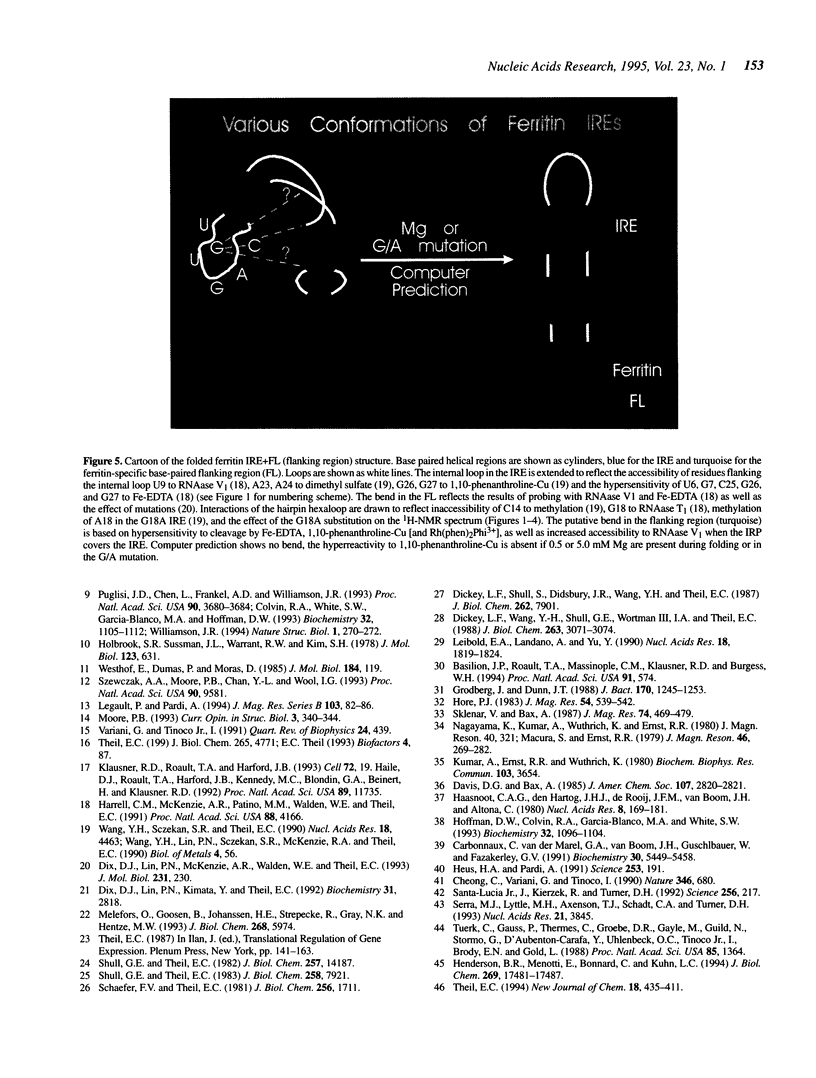
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Basilion J. P., Rouault T. A., Massinople C. M., Klausner R. D., Burgess W. H. The iron-responsive element-binding protein: localization of the RNA-binding site to the aconitase active-site cleft. Proc Natl Acad Sci U S A. 1994 Jan 18;91(2):574–578. doi: 10.1073/pnas.91.2.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonnaux C., van der Marel G. A., van Boom J. H., Guschlbauer W., Fazakerley G. V. Solution structure of an oncogenic DNA duplex containing a G.A mismatch. Biochemistry. 1991 Jun 4;30(22):5449–5458. doi: 10.1021/bi00236a018. [DOI] [PubMed] [Google Scholar]
- Cheong C., Varani G., Tinoco I., Jr Solution structure of an unusually stable RNA hairpin, 5'GGAC(UUCG)GUCC. Nature. 1990 Aug 16;346(6285):680–682. doi: 10.1038/346680a0. [DOI] [PubMed] [Google Scholar]
- Colvin R. A., White S. W., Garcia-Blanco M. A., Hoffman D. W. Structural features of an RNA containing the CUGGGA loop of the human immunodeficiency virus type 1 trans-activation response element. Biochemistry. 1993 Feb 2;32(4):1105–1112. doi: 10.1021/bi00055a016. [DOI] [PubMed] [Google Scholar]
- Dickey L. F., Sreedharan S., Theil E. C., Didsbury J. R., Wang Y. H., Kaufman R. E. Differences in the regulation of messenger RNA for housekeeping and specialized-cell ferritin. A comparison of three distinct ferritin complementary DNAs, the corresponding subunits, and identification of the first processed in amphibia. J Biol Chem. 1987 Jun 5;262(16):7901–7907. [PubMed] [Google Scholar]
- Dickey L. F., Wang Y. H., Shull G. E., Wortman I. A., 3rd, Theil E. C. The importance of the 3'-untranslated region in the translational control of ferritin mRNA. J Biol Chem. 1988 Mar 5;263(7):3071–3074. [PubMed] [Google Scholar]
- Dix D. J., Lin P. N., Kimata Y., Theil E. C. The iron regulatory region of ferritin mRNA is also a positive control element for iron-independent translation. Biochemistry. 1992 Mar 17;31(10):2818–2822. doi: 10.1021/bi00125a024. [DOI] [PubMed] [Google Scholar]
- Dix D. J., Lin P. N., McKenzie A. R., Walden W. E., Theil E. C. The influence of the base-paired flanking region on structure and function of the ferritin mRNA iron regulatory element. J Mol Biol. 1993 May 20;231(2):230–240. doi: 10.1006/jmbi.1993.1278. [DOI] [PubMed] [Google Scholar]
- Farmer B. T., 2nd, Müller L., Nikonowicz E. P., Pardi A. Unambiguous through-bond sugar-to-base correlations for purines in 13C,15N-labeled nucleic acids: the HsCsNb,HsCs(N)bCb, and HbNbCb experiments. J Biomol NMR. 1994 Jan;4(1):129–133. doi: 10.1007/BF00178341. [DOI] [PubMed] [Google Scholar]
- Grodberg J., Dunn J. J. ompT encodes the Escherichia coli outer membrane protease that cleaves T7 RNA polymerase during purification. J Bacteriol. 1988 Mar;170(3):1245–1253. doi: 10.1128/jb.170.3.1245-1253.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haasnoot C. A., den Hartog J. H., de Rooij J. F., van Boom J. H., Altona C. Loopstructures in synthetic oligodeoxynucleotides. Nucleic Acids Res. 1980 Jan 11;8(1):169–181. doi: 10.1093/nar/8.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haile D. J., Rouault T. A., Harford J. B., Kennedy M. C., Blondin G. A., Beinert H., Klausner R. D. Cellular regulation of the iron-responsive element binding protein: disassembly of the cubane iron-sulfur cluster results in high-affinity RNA binding. Proc Natl Acad Sci U S A. 1992 Dec 15;89(24):11735–11739. doi: 10.1073/pnas.89.24.11735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrell C. M., McKenzie A. R., Patino M. M., Walden W. E., Theil E. C. Ferritin mRNA: interactions of iron regulatory element with translational regulator protein P-90 and the effect on base-paired flanking regions. Proc Natl Acad Sci U S A. 1991 May 15;88(10):4166–4170. doi: 10.1073/pnas.88.10.4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson B. R., Menotti E., Bonnard C., Kühn L. C. Optimal sequence and structure of iron-responsive elements. Selection of RNA stem-loops with high affinity for iron regulatory factor. J Biol Chem. 1994 Jul 1;269(26):17481–17489. [PubMed] [Google Scholar]
- Heus H. A., Pardi A. Structural features that give rise to the unusual stability of RNA hairpins containing GNRA loops. Science. 1991 Jul 12;253(5016):191–194. doi: 10.1126/science.1712983. [DOI] [PubMed] [Google Scholar]
- Hoffman D. W., Colvin R. A., Garcia-Blanco M. A., White S. W. Structural features of the trans-activation response RNA element of equine infectious anemia virus. Biochemistry. 1993 Feb 2;32(4):1096–1104. doi: 10.1021/bi00055a015. [DOI] [PubMed] [Google Scholar]
- Holbrook S. R., Sussman J. L., Warrant R. W., Kim S. H. Crystal structure of yeast phenylalanine transfer RNA. II. Structural features and functional implications. J Mol Biol. 1978 Aug 25;123(4):631–660. doi: 10.1016/0022-2836(78)90210-3. [DOI] [PubMed] [Google Scholar]
- Jaeger J. A., Tinoco I., Jr An NMR study of the HIV-1 TAR element hairpin. Biochemistry. 1993 Nov 23;32(46):12522–12530. doi: 10.1021/bi00097a032. [DOI] [PubMed] [Google Scholar]
- Leclerc F., Cedergren R., Ellington A. D. A three-dimensional model of the Rev-binding element of HIV-1 derived from analyses of aptamers. Nat Struct Biol. 1994 May;1(5):293–300. doi: 10.1038/nsb0594-293. [DOI] [PubMed] [Google Scholar]
- Legault P., Pardi A. 31P chemical shift as a probe of structural motifs in RNA. J Magn Reson B. 1994 Jan;103(1):82–86. doi: 10.1006/jmrb.1994.1012. [DOI] [PubMed] [Google Scholar]
- Leibold E. A., Laudano A., Yu Y. Structural requirements of iron-responsive elements for binding of the protein involved in both transferrin receptor and ferritin mRNA post-transcriptional regulation. Nucleic Acids Res. 1990 Apr 11;18(7):1819–1824. doi: 10.1093/nar/18.7.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melefors O., Goossen B., Johansson H. E., Stripecke R., Gray N. K., Hentze M. W. Translational control of 5-aminolevulinate synthase mRNA by iron-responsive elements in erythroid cells. J Biol Chem. 1993 Mar 15;268(8):5974–5978. [PubMed] [Google Scholar]
- Michnicka M. J., Harper J. W., King G. C. Selective isotopic enrichment of synthetic RNA: application to the HIV-1 TAR element. Biochemistry. 1993 Jan 19;32(2):395–400. doi: 10.1021/bi00053a002. [DOI] [PubMed] [Google Scholar]
- Murphy F. L., Cech T. R. An independently folding domain of RNA tertiary structure within the Tetrahymena ribozyme. Biochemistry. 1993 May 25;32(20):5291–5300. doi: 10.1021/bi00071a003. [DOI] [PubMed] [Google Scholar]
- Murphy F. L., Cech T. R. GAAA tetraloop and conserved bulge stabilize tertiary structure of a group I intron domain. J Mol Biol. 1994 Feb 11;236(1):49–63. doi: 10.1006/jmbi.1994.1117. [DOI] [PubMed] [Google Scholar]
- Nikonowicz E. P., Pardi A. An efficient procedure for assignment of the proton, carbon and nitrogen resonances in 13C/15N labeled nucleic acids. J Mol Biol. 1993 Aug 20;232(4):1141–1156. doi: 10.1006/jmbi.1993.1466. [DOI] [PubMed] [Google Scholar]
- Puglisi J. D., Chen L., Frankel A. D., Williamson J. R. Role of RNA structure in arginine recognition of TAR RNA. Proc Natl Acad Sci U S A. 1993 Apr 15;90(8):3680–3684. doi: 10.1073/pnas.90.8.3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puglisi J. D., Tan R., Calnan B. J., Frankel A. D., Williamson J. R. Conformation of the TAR RNA-arginine complex by NMR spectroscopy. Science. 1992 Jul 3;257(5066):76–80. doi: 10.1126/science.1621097. [DOI] [PubMed] [Google Scholar]
- SantaLucia J., Jr, Kierzek R., Turner D. H. Context dependence of hydrogen bond free energy revealed by substitutions in an RNA hairpin. Science. 1992 Apr 10;256(5054):217–219. doi: 10.1126/science.1373521. [DOI] [PubMed] [Google Scholar]
- Schaefer F. V., Theil E. C. The effect of iron on the synthesis and amount of ferritin in red blood cells during ontogeny. J Biol Chem. 1981 Feb 25;256(4):1711–1715. [PubMed] [Google Scholar]
- Serra M. J., Lyttle M. H., Axenson T. J., Schadt C. A., Turner D. H. RNA hairpin loop stability depends on closing base pair. Nucleic Acids Res. 1993 Aug 11;21(16):3845–3849. doi: 10.1093/nar/21.16.3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shull G. E., Theil E. C. Regulation of ferritin mRNA: a possible gene-sparing phenomenon. Induction of ferritin synthesis by iron in liver as well as red cells combines high translational efficiency with increased utilization of preformed ferritin mRNA. J Biol Chem. 1983 Jul 10;258(13):7921–7923. [PubMed] [Google Scholar]
- Shull G. E., Theil E. C. Translational control of ferritin synthesis by iron in embryonic reticulocytes of the bullfrog. J Biol Chem. 1982 Dec 10;257(23):14187–14191. [PubMed] [Google Scholar]
- Sklenár V., Peterson R. D., Rejante M. R., Feigon J. Correlation of nucleotide base and sugar protons in a 15N-labeled HIV-1 RNA oligonucleotide by 1H-15N HSQC experiments. J Biomol NMR. 1994 Jan;4(1):117–122. doi: 10.1007/BF00178339. [DOI] [PubMed] [Google Scholar]
- Szewczak A. A., Moore P. B., Chang Y. L., Wool I. G. The conformation of the sarcin/ricin loop from 28S ribosomal RNA. Proc Natl Acad Sci U S A. 1993 Oct 15;90(20):9581–9585. doi: 10.1073/pnas.90.20.9581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theil E. C. Regulation of ferritin and transferrin receptor mRNAs. J Biol Chem. 1990 Mar 25;265(9):4771–4774. [PubMed] [Google Scholar]
- Tuerk C., Gauss P., Thermes C., Groebe D. R., Gayle M., Guild N., Stormo G., d'Aubenton-Carafa Y., Uhlenbeck O. C., Tinoco I., Jr CUUCGG hairpins: extraordinarily stable RNA secondary structures associated with various biochemical processes. Proc Natl Acad Sci U S A. 1988 Mar;85(5):1364–1368. doi: 10.1073/pnas.85.5.1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhof E., Dumas P., Moras D. Crystallographic refinement of yeast aspartic acid transfer RNA. J Mol Biol. 1985 Jul 5;184(1):119–145. doi: 10.1016/0022-2836(85)90048-8. [DOI] [PubMed] [Google Scholar]
- Wijmenga S. S., Heus H. A., Werten B., van der Marel G. A., van Boom J. H., Hilbers C. W. Assignment strategies and analysis of cross-peak patterns and intensities in the three-dimensional homonuclear TOCSY-NOESY of RNA. J Magn Reson B. 1994 Feb;103(2):134–141. doi: 10.1006/jmrb.1994.1021. [DOI] [PubMed] [Google Scholar]
- Williamson J. R. RNA origami. Nat Struct Biol. 1994 May;1(5):270–272. doi: 10.1038/nsb0594-270. [DOI] [PubMed] [Google Scholar]