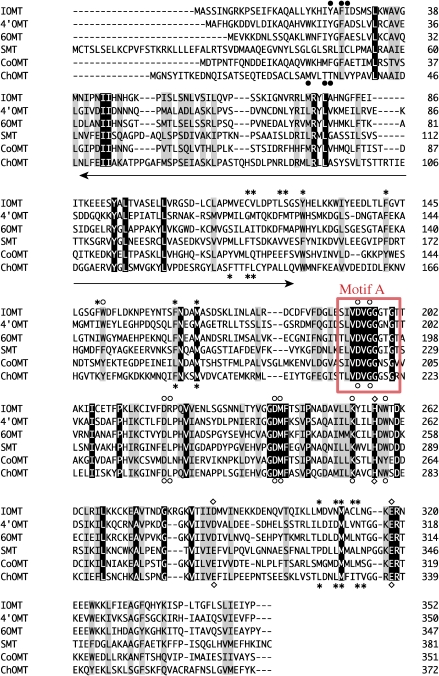
Figure 2.
Amino acid sequence alignment of Coptis and flavonoid OMTs. The substrate-binding residues (asterisk), AdoMet-binding residues (open circle), catalytic residues (square) and substrate-binding from the partner monomer (solid circle) are shown for S-adenosyl-L-methionine:isoflavone O-methyltransferase (IOMT) and S-adenosyl-L-methionine:chalcone O-methyltransferase (ChOMT).11) Amino acids conserved in all of the sequences are highlighted, and similar ones are shaded. The arrow indicates the 90-amino acid region that is believed to be important for alkaloid recognition. Motif A is boxed in red.