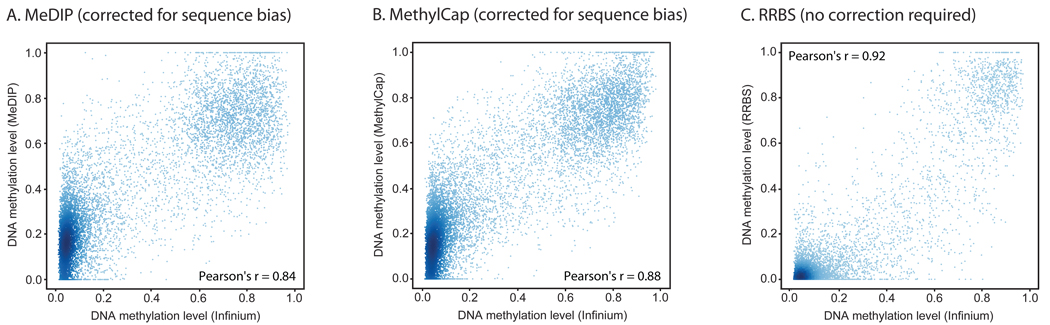
Figure 3. Quantification of DNA methylation with MeDIP, MethylCap and RRBS.
Absolute DNA methylation levels were calculated from the data obtained by MeDIP (panel A), MethylCap (panel B) and RRBS (panel C), respectively, and compared to DNA methylation levels determined by the Infinium assay. For MeDIP and MethylCap, sequencing reads were counted in 1-kilobase regions surrounding each CpG that is interrogated by the Infinium assay, and a regression model was used to infer absolute DNA methylation levels. Scatterplots and correlation coefficients were calculated on a test set that was not used for model fitting or feature selection. For RRBS, the DNA methylation level was determined as the percentage of methylated CpGs within 200 basepairs surrounding each CpG that is interrogated by the Infinium assay. Data shown are for the HUES6 human ES cell line, and regions that did not have sufficient sequencing coverage were excluded.