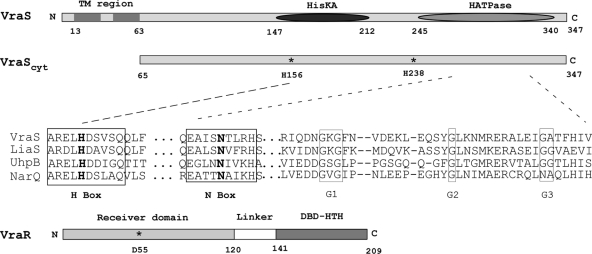
FIG. 1.
Schematic diagram of VraS and VraR highlighting their domain architecture and the location of amino acids mutated in the present study. The predicted transmembrane region (TM), histidine kinase (HisKA), and histidine kinase ATP-binding domain (HATPase) are shown. VraScyt indicates the point of truncation used for the purification of the cytoplasmic and soluble portion of the protein used in the present study. The VraR response regulator is depicted and shows the receiver domain and position of phosphorylated aspartate, along with the C-terminal DNA-binding domain. The sequence alignment shows the H-box sequence context of VraS-H156, together with other conserved motifs within the ATP-binding domain. Sequences used were from Swiss-Prot accession: Staphylococcus aureus VraS, Q99SZ7; Bacillus subtilis LiaS, O32198; Escherichia coli UhpB, P09835; and Haemophilus influenzae NarQ, P44604.