Abstract
The pyrrole polyether antibiotic calcimycin (A23187) is a rare ionophore that is specific for divalent cations. It is widely used as a biochemical and pharmacological tool because of its multiple, unique biological effects. Here we report on the cloning, sequencing, and mutational analysis of the 64-kb biosynthetic gene cluster from Streptomyces chartreusis NRRL 3882. Gene replacements confirmed the identity of the gene cluster, and in silico analysis of the DNA sequence revealed 27 potential genes, including 3 genes for the biosynthesis of the α-ketopyrrole moiety, 5 genes that encode modular type I polyketide synthases for the biosynthesis of the spiroketal ring, 4 genes for the biosynthesis of 3-hydroxyanthranilic acid, an N-methyltransferase tailoring gene, a resistance gene, a type II thioesterase gene, 3 regulatory genes, 4 genes with other functions, and 5 genes of unknown function. We propose a pathway for the biosynthesis of calcimycin and assign the genes to the biosynthesis steps. Our findings set the stage for producing much desired calcimycin derivatives using genetic modification instead of chemical synthesis.
Calcimycin (A23187) is one of few natural ionophore antibiotics that specifically transport divalent cations such as calcium and magnesium (25). Calcimycin inhibits the growth of Gram-positive bacteria and some fungi (20). It also inhibits the activity of ATPase and uncouples oxidative phosphorylation of mammalian cells, and it induces apoptosis of cultured cells via direct activation of intracellular signals (17, 29). Calcimycin is widely used as a biochemical tool for pharmacological and in vitro toxicological studies, and it has been mentioned in more than 16,000 publications since its discovery in 1972.
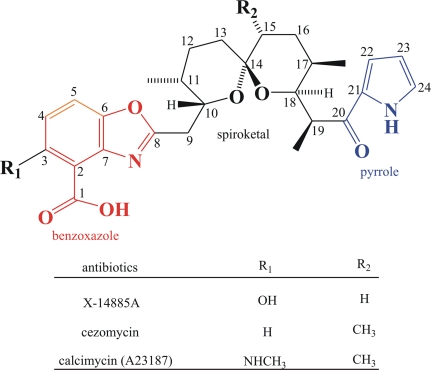
There are two additional natural pyrrole polyether antibiotics similar in structure to calcimycin: X-14885A from Streptomyces chartreusis (NRRL 12350) and cezomycin (8, 19, 35). Cezomycin is also produced by Streptomyces chartreusis NRRL 3882 as a precursor for the much more valuable calcimycin (8). All three antibiotics feature an α-ketopyrrole, a substituted benzoxazole, and a spiroketal ring. They differ only in the nature of two side groups (Fig. 1).
FIG. 1.
Structures of calcimycin, cezomycin, and X-14885A.
Calcimycin and a number of similar molecules have been chemically synthesized (26-28). Calcimycin derivatives have also been generated by microbial transformation (1). It would, however, be desirable to produce additional calcimycin derivatives by fermentation using genetically modified bacteria. This requires targeted mutation of the sequenced antibiotic biosynthesis gene cluster.
The cloning of the calcimycin biosynthetic genes was greatly aided by the data from feeding experiment using isotopes and nuclear magnetic resonance analysis, which indicated that the α-ketopyrrole is derived from l-proline, the spiroketal ring is a polyketide derived from propionate and acetate, the benzoxazole is synthesized from glucose via a shikimate-type pathway that produces 3-hydroxyanthranilic acid, and the N-methyl group of the benzoxazole comes from methionine (7, 39).
Here we report on the cloning and sequencing of a 64-kb gene cluster that contains genes for the synthesis of the three parts of the calcimycin backbone (benzoxazole, spiroketal ring, and α-ketopyrrole), a modification gene, a resistance gene (to prevent the killing of the producer strain), a type II thioesterase gene, three regulatory genes, four genes with other functions, and five genes of unknown function.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli was grown at 37°C in liquid Luria-Bertani medium or on solid Luria-Bertani medium containing 2% agar, as described by Sambrook and Russell (31). S. chartreusis NRRL 3882 was cultured at 30°C in tryptic soy broth and yeast extract (TSBY) medium for the isolation of chromosomal DNA and on solid mannitol soya flour (SFM) medium for spore collection and conjugation (15).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Reference or source |
|---|---|---|
| Strains | ||
| Streptomyces chartreusis | ||
| NRRL 3882 | Wide type, calcimycin producer | NRRL |
| WQL1 | ΔcalN2, calcimycin negative | This work |
| WQL2 | ΔcalN1, calcimycin negative | This work |
| WQL4 | ΔcalB2, calcimycin negative | This work |
| WQL5 | ΔcalN2::calN2, calcimycin producer | This work |
| Escherichia coli | ||
| DH10B | F−recA lacZΔM15 | 6 |
| ET12567(pUZ8002) | Cml Kan dam dcm hsdSB Tra+ Cml | 15 |
| BW25113(pIJ790) | araBp-gam-be-exo AraC [RepA101(Ts) Cml] | 12 |
| BL21(DE3)/plysE | F−dcm ompT hsdSB(r-B− mB−) gal λ(DE3)(pLysE Cml) | Stratagene |
| EPI300-T1R/pCC1FOS | CopyControl fosmid library production kit | EPICENTRE |
| Plasmids | ||
| pBluescript II SK(+) | bla lacZ orif1 | Stratagene |
| pOJ446 | aac(3)IV ori (SCP2*) reppucoriTcos | 3 |
| pSET152 | aac(3)IV lacZ reppucattΦC31 oriT | 3 |
| pMD18-T | pUC18 derivative T vector | TaKaRa |
| pJTU3157 | pBluescript II SK(+)-derived plasmid carrying calN1 and calB2 | This work |
| pJTU3167 | pOJ446-derived plasmid carrying calN2 | This work |
| pJTU3170 | pJTU3167-derived plasmid, neo, calN2-defective plasmid | This work |
| pJTU3171 | pOJ446-derived plasmid carrying calN1 and calB2 | This work |
| pJTU3175 | pJTU3171-derived plasmid carrying neo resistance gene, calN1-defective plasmid | This work |
| pJTU3178 | pJTU3171-derived plasmid carrying neo resistance gene, calB2-defective plasmid | This work |
| pJTU3185 | pSET152-derived plasmid carrying calN2, ΔcalN2 complementation plasmid | This work |
Cml, chloramphenicol resistance; Kan, kanamycin resistance; aac(3)IV, apramycin resistance; neo, kanamycin resistance.
The antibiotics ampicillin (100 μg/ml), apramycin (30 μg/ml), chloramphenicol (25 μg/ml), and kanamycin (50 μg/ml) were used for the selection of recombinants.
DNA isolation, manipulation, and plasmid construction.
DNA isolation and manipulation were performed as described by Sambrook and Russell (31). DNA fragments were recovered from agarose gels using a Tiangen purification kit (Beijing, China), according to the supplied protocol. Restriction endonucleases and other enzymes were purchased from NEB Co. Ltd. Enzyme reactions were carried out according to the instructions provided by the manufacturer. Synthetic primers were purchased from Sangon Corp. (Shanghai, China). Clones were sequenced by Invitrogen Corp. (Shanghai, China).
Cloning and sequencing of calcimycin biosynthetic gene cluster. (i) PCR amplification of the proline adenyltransferase gene from S. chartreusis NRRL 3882.
Fifteen sets of degenerate primers were designed on the basis of the conserved domains of bacterial proline adenytransferase genes found in pyoluteorin, coumermycin A1, prodiginines, clorobiocin, and pyrrolomycin biosynthetic gene clusters (5, 23, 24, 34, 37). PCR amplification was run for 30 cycles. The conditions for each cycle were 30 s at 94°C, 30 s at 63°C, and 1 min at 72°C. Four different PCR fragments were purified after agarose gel separation and cloned into the PMD 18-T vector (TaKaRa Corp.) for sequencing. One pair of successful primers was QL1-FP1 and QL1-RP3 (Table 2).
TABLE 2.
Primers used in this study
| Primer | Oligonucleotide sequencea |
|---|---|
| QL1-FP1 | 5′-CTC TAC ACC TCC GGN TCS ACC GG-3′ |
| QL1-RP3 | 5′-GGT GCA GAC GTT GGT YTC SGT RGG NCC-3′ |
| QL2-FP1 | 5′-TAGGGCCGTAGAGGTTCAGCA-3′ |
| QL2-RP1 | 5′-GGGGTCTGCATCAGCCACC-3′ |
| QL7-FP1 | 5′-TGGTGAAGGACGGGGAGG-3′ |
| QL7-RP1 | 5′-CGAGCGGGAGGAACGACA-3′ |
| QL7-FP3 | 5′-AGGATGCCCAGCACGACCAC-3′ |
| QL7-RP3 | 5′-CCCGATTCACGGACCTCAACT-3′ |
N is A, T, G, or C; S is G or C; Y is C or T; and R is A or G.
(ii) Construction and screening of a cosmid library.
The S. chartreusis NRRL 3882 cosmid library was constructed using a CopyControl fosmid library production kit, according to the protocol provided by the company. A pair of nondegenerate QL2P1 primers (QL2-FP1 and QL2-RP1) were designed according to the above sequencing result, and the labeled PCR product was then used to screen the cosmid library (Fig. 2 and 3; Table 2).
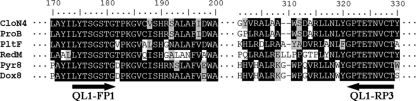
FIG. 2.
Alignment of bacterial proline adenyltransferase sequences used to design degenerate primers for the amplification of heterologous proline adenytransferase genes: CloN4 (Streptomyces roseochromogenes DS 12.976), ProB (Streptomyces rishiriensis DSM 40489), PltF (Pseudomonas fluorescens Pf-5), RedM [Streptomyces coelicolor A3(2)], Pyr8 (Actinosporangium vitaminophilum ATCC 31673), and Dox8 (Streptomyces sp. strain UC 11065). The degenerate primers QL1-FP1 and QL1-RP3 were designed according to the highly conserved amino acid sequence indicated by black arrows, taking into account the biased codon usage of the high G+C content of Streptomyces DNA.
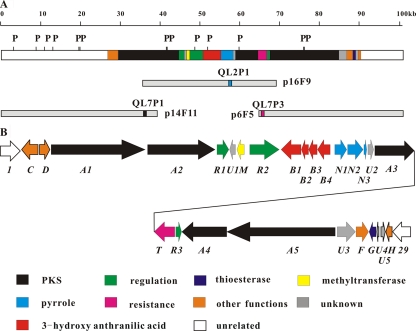
FIG. 3.
Calcimycin biosynthesis gene cluster from S. chartreusis NRRL 3882. (A) PstI (P) restriction map of the 101-kb region covered by cosmids p14F11, p16F9, and p6F5. Filled areas indicate the 64 kb of DNA represented in panel B. The gray rectangles indicate the extent of the cosmids. QL2P1, QL7P1, and QL7P3 are labeled PCR fragments that were used as probes to screen the cosmid library. (B) Open reading frames of the calcimycin biosynthetic gene cluster. The prefix cal was omitted from the gene designations. White ORFs 1 and 29 are probably not involved in calcimycin biosynthesis. The proposed functions for the individual ORFs are summarized in Table 3.
PCR primers QL7P1 (QL7-FP1 and QL7-RP1) and QL7P3 (QL7-FP3 and QL7-RP3) were then designed to amplify sequences near the two ends of cosmid p16F9 (Table 2). The labeled PCR products were then used to identify flanking cosmids, such as p6F5 and p14F11 (Fig. 3). Sets of cosmids that hybridized to one of the PCR probes were digested using PstI. The large numbers of cosmid digests that were compared made it possible to deduce the PstI restriction map of the entire contig. This, in turn, made it possible to select the two cosmids p6F5 and p14F11 that overlapped the central cosmid p16F9 only a short distance.
The three cosmids were sequenced, and the sequences were aligned using BioEdit software, version 4.8.10. Open reading frames (ORFs) were predicted using FramePlot software, version 4.0 (http://nocardia.nih.go.jp/fp4/). The putative proteins were compared with data online using the BLAST program (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Module and domain organizations of polyketide synthases (PKSs) were deduced by NRPS-PKS software online (http://www.nii.res.in/nrps-pks.html) (2, 36).
Gene inactivation and complementation.
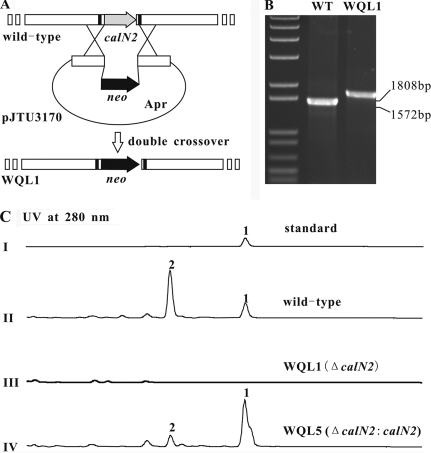
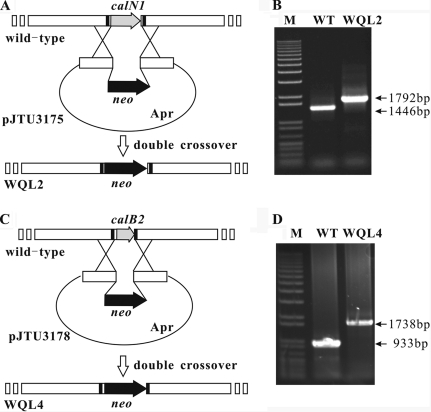
The targeted genes from the calcimycin gene cluster of S. chartreusis NRRL 3882 were disrupted using the Redirct technology, according to the supplied protocol, with some modifications (12). Briefly, the neo gene from SuperCos1 was used to replace an internal region of the target gene. For the construction of the calN2 disruption plasmid, a 3,679-bp BamHI fragment encoding calN2 from cosmid p16F9 was cloned into the BamHI site of pOJ446 to produce pJTU3167. For the construction of the calN1 and calB2 disruption plasmids, a 7,889-bp PstI fragment harboring calN1 and calB2 from cosmid p16F9 was cloned into the PstI site of pBluescript SK(+), generating pJTU3157. The 7,889-bp SpeI/EcoRV fragment from pJTU3157 was cloned into the SpeI/EcoRV sites of pOJ446 to yield pJTU3171. The neo gene, amplified using KOD-plus DNA polymerase (Toyobo), was introduced into pJTU3167 and pJTU3171 using the PCR targeted method, generating the desired plasmids, pJTU3170 (ΔcalN2), pJTU3175 (ΔcalN1), and pJTU3178 (ΔcalB2). These mutant plasmids were then introduced into S. chartreusis NRRL 3882 by conjugation from E. coli ET12567/pUZ8002. Apramycin-sensitive and kanamycin-resistant colonies were isolated as WQL1 (ΔcalN2), WQL2 (ΔcalN1), and WQL4 (ΔcalB2). Successful gene disruptions were confirmed by PCR and high-pressure liquid chromatography (HPLC) and mass spectrometric (MS) analysis (Fig. 4 to 6).
FIG. 4.
Construction and analysis of a calN2 (putative proline adenyltransferase) mutant strain. (A) Double-crossover replacement to generate strain WQL1. The black rectangles indicate the positions of the primers used to generate the PCR products shown in panel B. (B) Ethidium bromide-stained agarose gel. The formation of a larger band from WQL1 confirmed the gene replacement. WT, wild type. (C) HPLC analysis of calcimycin production in S. chartreusis NRRL 3882. I, calcimycin standard purchased from Sigma-Aldrich; II, extract from wild-type S. chartreusis NRRL 3882; III, mutant WQL1 (ΔcalN2); IV, complemented strain WQL5 (ΔcalN2::calN2). The functional copy of calN2 was cloned on the integrating vector pSET152. Compounds: 1, calcimycin; 2, cezomycin (Fig. 1). Note that the relative amounts of compounds 1 and 2 produced by the wild type and WQL5 varied considerably in repeated experiments.
FIG. 5.
Construction of strains WQL2 and WQL4 by double-crossover deletion of the genes calN1 and calB2, respectively. (A) Generation of WQL2; (B) PCR amplification using flanking primers (black rectangles) confirmed the replacement of calN1 by neo; (C) generation of WQL4; (D) PCR amplification using flanking primers (black rectangles) confirmed the replacement of calB2 by neo. WT, wild type.
FIG. 6.
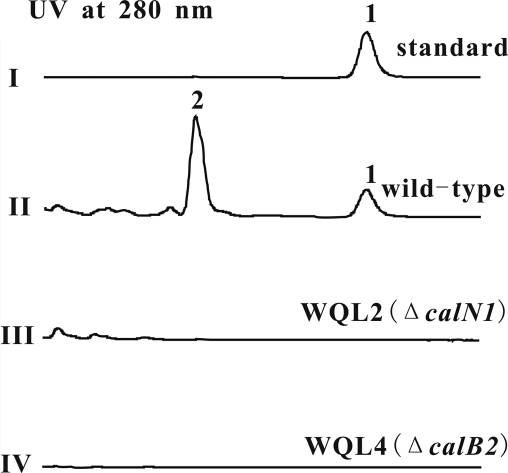
HPLC analysis showing the lack of calcimycin production by S. chartreusis mutant strains WQL2 (ΔcalN1) and WQL4 (ΔcalB2). I, calcimycin standard (Sigma-Aldrich); II, S. chartreusis NRRL 3882 wild type producing calcimycin (1) and cezomycin (2); III, nonproducing mutant WQL2 (ΔcalN1); IV, nonproducing mutant WQL4 (ΔcalB2). See Fig. 1 for the structures of calcimycin and cezomycin.
To construct the plasmid for gene complementation of calN2, a BamHI-digested fragment carrying calN2 (from pJTU3166) was cloned into the BamHI site of pSET152, generating pJTU3185. Introduction of pJTU3185 into WQL1 (ΔcalN2) via conjugation generated the complemented strain, WQL2.
HPLC and MS analysis of calcimycin biosynthesis.
S. chartreusis NRRL 3882 and its mutant derivatives were cultivated at 30°C for 6 to 10 days with SFM medium. Then, the culture medium was extracted with an equal volume of methanol. The resulting extracts were concentrated in vacuo and redissolved in methanol for HPLC/MS analysis using an Agilent 1100 series LC/MSD Trap system and an Agilent Zorbax SB-C18 (2.1 by 50 mm) column. HPLC was performed with a linear gradient of 60% of 85% CH3OH-H2O (0.1% trifluoroacetic acid) over 30 min at a flow rate of 0.2 ml/min and monitoring at 280 nm. The calcimycin standard, purchased from Sigma-Aldrich, was used as a control.
Nucleotide sequence accession number.
The nucleotide sequence obtained in this study was submitted to GenBank database, under accession no. HM452329.
RESULTS
Cloning and sequencing of calcimycin biosynthetic gene cluster.
Calcimycin contains three distinctive structures: a spiroketal ring, a benzoxazole heterocycle, and an α-ketopyrrole moiety (Fig. 1). Of these, only α-ketopyrrole moieties are found in a wide variety of biologically active natural products. Feeding experiments have shown that the α-ketopyrrole of calcimycin is derived from l-proline (11, 33). Biosynthesis of α-ketopyrrole starts with the activation of l-proline to l-prolyl-AMP by a specific adenyltransfease. Aligning the amino acid sequences of six adenyltransferases involved in the biosynthesis of five different antibiotics identified four conserved regions (Fig. 2). We synthesized degenerate primers according to these conserved sequences, and using S. chartreusis genomic DNA as a template, we obtained a PCR fragment of the expected size of 460 bp. The sequence of the DNA fragment was consistent with it encoding a S. chartreusis l-proline adenyltransferase (CalN2 in Fig. 3). We then used the labeled 460-bp DNA fragment as a hybridization probe to identify S. chartreusis cosmid clones that contained parts of the calcimycin biosynthesis gene cluster.
Several hybridizing cosmids were isolated and positioned relative to each other according to their PstI restriction maps (Fig. 3). All the cosmids formed a single ca. 64-kb contig. One of the central cosmids, p16F9, was sequenced. It contained on both ends incomplete type I PKS genes which were likely to be involved in the biosynthesis of calcimycin.
To find flanking cosmids that overlapped p16F9 only by a short sequence, PCR probes QL7P1 and QL7P3 (Fig. 3A) were generated from either end for chromosome walking. PstI restriction mapping identified cosmids p6F5 and p14F11, which extend far on either side of p16F9. Together, the three contiguous cosmids cover a 101-kb region with an overall G+C content of 72.0%, which is normal for Streptomyces (the sequence of the 64-kb region shown schematically in Fig. 3B has been deposited in GenBank under accession no. HM452329).
Translated open reading frames were identified using FramePlot, and database searches identified 27 clustered ORFs spanning a 64-kb DNA region. We chose the designation cal (calcimycin) for all these putative antibiotic biosynthesis genes (Fig. 3B; Table 3) .
TABLE 3.
Deduced functions of ORFs in the calcimycin biosynthetic gene cluster
| Genea | Sizeb | Putative function | Homologc | % identity/% similarity |
|---|---|---|---|---|
| cal1 | 680 | Peptidase S15 | SvirD4_04762 (ZP_05529758) | 94/97 |
| calC | 521 | Fatty acid-CoA ligase | FadD (CAM60387) | 63/77 |
| calD | 331 | Oxidoreductase | SvirD4_04782 (ZP_05529762) | 92/94 |
| calA1 | 2,942 | PKS | ||
| calA2 | 2,063 | PKS | ||
| calR1 | 398 | TylR family regulator | TylR (AAF29380) | 39/51 |
| calU1 | 183 | Unknown | SvirD4_04827 (ZP_05529769) | 92/97 |
| calM | 229 | N-Methyltransferase | AtaP5 (CAD27648) | 42/56 |
| calR2 | 1,040 | LuxR family regulator | LuxR (ABP55203) | 35/49 |
| calB1 | 534 | Anthranilate synthase | PhzE (AAF17499) | 57/74 |
| calB2 | 226 | Isochorismatase | PhzD (AAF17498) | 63/72 |
| calB3 | 270 | 2,3-Dihydroxybenzoate-2,3-dehydrogenase | DhbA (BAI86724) | 57/74 |
| calB4 | 404 | 3-Deoxy-d-arabino-heptulosonate 7-phosphate synthase | PhzC (AAF17497) | 54/68 |
| calN1 | 379 | Acyl-CoA dehydrogenase | CloN3 (AAN65232) | 61/73 |
| calN2 | 495 | Adenylation for l-proline | CloN4 (AAN65233) | 60/70 |
| calN3 | 87 | Proline carrier protein | CloN5 (AAN65234) | 48/72 |
| calU2 | 180 | Unknown | Aln8 (ACI88878) | 29/46 |
| calA3 | 1,618 | PKS | ||
| calT | 749 | Transmembrane transport protein | AlnT2 (ACI88887) | 51/67 |
| calR3 | 211 | TetR family regulator | MonRII (AAO65794) | 57/75 |
| calA4 | 1,638 | PKS | ||
| calA5 | 3,956 | PKS | ||
| calU3 | 661 | Unknown | SvirD4_04937 (ZP_05529789) | 92/95 |
| calF | 445 | 3-Carboxymuconate cycloisomerase | SvirD4_04942 (ZP_05529790) | 87/90 |
| calG | 251 | Type II thioesterase | IdmA (ACN69977) | 59/73 |
| calU4 | 72 | Unknown | SvirD4_04952 (ZP_05529792) | 90/94 |
| calU5 | 154 | Unknown | SvirD4_04957 (ZP_05529793) | 94/97 |
| calH | 212 | Chorismate mutase | SCAB_58971 (YP_003491457) | 64/70 |
| cal29 | 460 | Peptidase M20 | BgramDRAFT_0002 (ZP_02881193) | 67/79 |
In order of sequence.
Number of amino acid residues.
Designations in parentheses are NCBI accession numbers.
Deletion of calN1, calN2, and calB2 proved that these genes are required for calcimycin biosynthesis.
We predicted that calN1 (acyl coenzyme A [acyl-CoA] dehydrogenase) and calN2 (l-proline adenyltransferase) are required for the biosynthesis of the α-ketopyrrole structure and that calB2, isochorismatase, may be required for the synthesis of the 3-hydroxyanthranilic acid moiety. The three genes were individually deleted from the calcimycin producer using gene replacements, as shown in Fig. 4 to 6. The resulting mutant strains were tested for antibiotic production using HPLC/MS. As expected, deletion of each of these three genes abolished the production of calcimycin.
To make sure that the lack of antibiotic production was indeed caused by the gene replacement rather than by a spontaneous mutation elsewhere in the S. chartreusis genome, we reintroduced calN2 cloned onto a plasmid, pSET152, into the strain from which this gene had been removed. This trans complementation restored calcimycin production and thus proved that calN2 is required for calcimycin production (Fig. 4).
In silico analysis of the calcimycin gene cluster. (i) Pyrrole moiety biosynthesis.
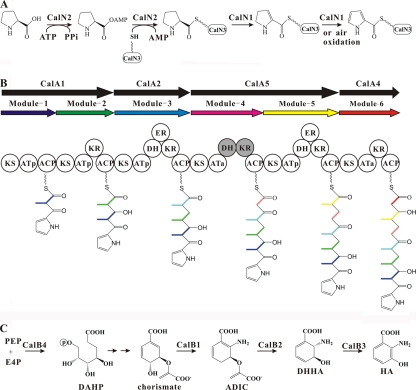
The putative proteins CalN1 to CalN3 are most similar to CloN3 to CloN5, respectively (61%, 60%, and 48% amino acid identities, respectively), of Streptomyces roseochromogenes DS 12.976, which have been proved to activate l-proline by converting it to a thioester of pyrrole-2-carboxylic acid (24). CalN2 first adenylates proline and then replaces the adenyl group with CalN3, forming a thioester bond. CalN1 oxidizes the prolyl ring to create α-ketopyrrole. The second double bond might form spontaneously by air oxidation. The α-ketopyrrole on CalN3 is then used as a primer for polyketide synthesis (Fig. 7A).
FIG. 7.
Proposed pathways for the biosynthesis of the pyrrole, polyketide, and benzoxazole moieties of calcimycin. (A) CalN2 adenylates l-proline and then replaces the adenyl group with CalN3SH. CalN1 creates two double bonds in the prolyl ring to create the α-ketopyrrole derivative. The second double bond could also form spontaneously by air oxidation. (B) The modular PKS assembly line uses α-ketopyrrole-S-CalN3 as the starter unit and four methylmalonyl and two malonyl units to generate the polyketide chain of calcimycin. The black arrows indicate multidomain PKS synthases containing one or two PKS modules. The circles indicate catalytic centers (domains): KS, AT (p, methylmanlonyl-CoA specific; a, malonyl-CoA specific), DH, KR, ER, and ACP. The shaded circles in module 4 indicate that the DH or KR activity has been lost by point mutations. Note that CalA3 is missing from this scheme because it was predicted to be enzymatically inactive (Table 4). (C) Synthesis of 3-hydroxyanthranilic acid catalyzed by CalB1 to CalB4. Arrows without an enzyme designation indicate reactions that are not predicted to be catalyzed by any of the genes shown in Fig. 3. Putative functions of the genes: CalB1, anthranilate synthase; CalB2, isochorismatase; CalB3, 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase; CalB4, 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase. Compounds: PEP, phosphoenolpyruvate; E4P, erythrose-4-phosphate; DAHP, 3-deoxy-d-arabino-heptulosonate 7-phosphate; ADIC, 2-amino-2-deoxyisochorismate; DHHA, trans-2,3-dihydro-3-hydroxyanthranilic acid; HA, 3-hydroxyanthranilic acid.
(ii) Polyketide chain biosynthesis.
The noncontiguous genes calA1 to calA5 encode five noncontiguous type I multifunctional, modular PKSs (Fig. 3B; Table 3).
Each PKS module should minimally contain a ketosynthase (KS), an acyltransferase (AT), and an acyl carrier protein (ACP). Optionally, there are also a dehydratase (DH), an enoyl reductase (ER), and a β-ketoreductase (KR). Missing optional activities may have inactivating point mutations, or the entire domain may be deleted. The optional domains determine the reduction state of the incorporated extender unit, which can be a keto group, a hydroxyl group, a trans double bond, or a saturated CH2.
The NRPS-PKS online software was used to deduce the functionalities of the five CalA PKSs containing seven PKS modules (Table 4) (2, 36). Each module has a different predicted functionality which made it easy to assign the modules unambiguously to the sequence of polyketide synthesis predicted for calcimycin (Fig. 8, stgructure 3, which shows the extender units in boldface). Note that CalA3 is not predicted to be involved in polyketide chain elongation because it has an inactive AT. (The missing ACP could possibly be provided in trans from a type II PKS or fatty acid synthetase.) The pyrrole-2-carboxylic acid starter unit is transferred from CalN3 to the active-site cysteine residue of the KS domain of module 1 to prime the biosynthesis of the polyketide chain. Finally, CalG resembles type II thioesterases (TEs) and probably releases the completed polyketide from the PKS, and it may also serve as an editing enzyme by cleaving miscognate chains from the ACP domains (9, 13, 16, 38).
TABLE 4.
PKSs organization in the calcimycin biosynthetic gene cluster
| CalA module | Order of use | PKS domain in the order of the sequencea |
Predicted functionalityf | |||||
|---|---|---|---|---|---|---|---|---|
| KS (3) | ATb (1) | DHc (5) | ERd (6) | KRe (4) | ACP (2) | |||
| 1a | 1 | 1 | 1p | Not present | Not present | Not present | 1 | C=O, CH3 |
| 1b | 2 | 2 | 2p | Not present | Not present | 2d (C-18) K-S-Y | 2 | CHOH, CH3 |
| 2 | 3 | 3 | 3p | 3 HXXXGXXXXP | 3 | 3 K-S-Y | 3 | CH2, CH3 |
| 5a | 4 | 4 | 4a | Inactive, HXXXEXXXXP | Not present | Inactive, R-S-L | 4 | C=O |
| 5b | 5 | 5 | 5p | 5 HXXXGXXXXP | 5 | 5 K-S-Y | 5 | CH2, CH3 |
| 4 | 6 | 6 | 6a | Not present | Not present | 6l (C-10) K-S-Y | 6 | CHOH |
| 3 | ? | Present | Inactive, S → G | Not present | Not present | Inactive, P-S-G | Not present | No chain elongation |
A bold number means that the indicated functional domain is predicted to be active. The numbers in parentheses in the subheads refer to those in the order-of-use column.
a, malonate is used to incorporate acetate; p, methylmalonate is used to incorporate propionate. Active AT domains feature the conserved catalytic key residue Ser.
Functional DH domains have the conserved motif HXXXGXXXXP. The mutated conserved amino acids are printed in boldface.
Functional ER domains feature the conserved motif GXGXAAXXXA.
Functional KR domains contain the conserved consensus motif GXGXXGXXXA associated with the NADP(H) binding site and the K-S-Y catalytic triad. The letters d and l after the module number indicate the predicted stereospecific configuration at the β position of chiral centers, as described previously (30). Note that a chiral center is formed only when the KR is active and the ER and DH are inactive or missing. The number of the C atom is indicated in parentheses, as in Fig. 1. In CalA3, the KR GXGXXGXXXA motif is changed to GXTXXAXXXL. The mutated conserved amino acids are printed in boldface.
Redox state of the β-carbon atom; CH3, propionate instead of acetate.
FIG. 8.
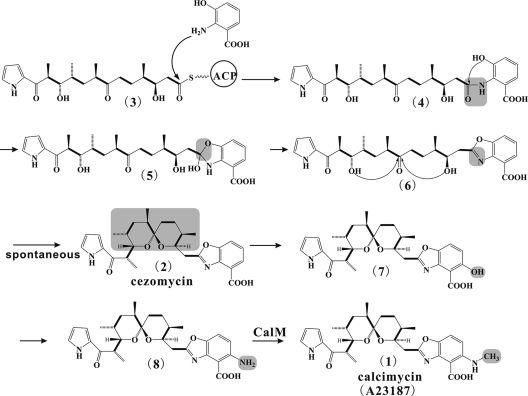
Proposed pathway for the generation of the benzoxazole ring and further maturation of the calcimycin molecule. The amino group of 3-hydroxyanthranilic acid nucleophilically attacks the terminal carbonyl group of the polyketide chain (substance 3) and releases the polyketide chain (substance 4) from the ACP of the PKS module 6 (Fig. 7). An acetalization reaction then closes the heterocycle ring and generates the benzoxazole moiety (substance 5). Spiroketal ring formation (structure 6) may proceed spontaneously to give cezomycin (substance 2). Hydroxylation (substance 7), amination (substance 8), and N-methylation (catalyzed by CalM) finally generate calcimycin (substance 1). Note that only substances 1 and 2 have actually been observed. The gray background indicates where the last change in the reaction series has taken place. CalD and CalG are only tentatively assigned to the respective reactions.
(iii) 3-Hydroxyanthranilic acid biosynthesis.
Four genes (calB1 through calB4) within the cal gene cluster encode proteins which are very similar to known genes that are involved in the biosynthesis of 3-hydroxyanthranilic acid prior to attachment to the finished polyketide chain (Fig. 8, structure 3 to give structure 4) (22). The proposed gene assignment for the pathway is shown in Fig. 7C and Table 3.
(iv) Tailoring, regulation, resistance, and other functions.
CalM resembles S-adenosylmethionine (SAM)-specific N-methyltransferases (32). The function of CalM was recently confirmed by gene replacement in vivo and biochemical studies in vitro (Q. Wu, submitted for publication). CalR1 resembles TylR family regulatory proteins, CalR2 resembles LuxR family regulatory proteins, and CalR3 resembles TetR-type transcriptional regulators. Because of their location within the calcimycin gene cluster, we predict that all three genes are likely to participate in the regulation of calcimycin biosynthesis. CalT resemble transmembrane proteins and could be responsible for the export of calcimycin into the medium and rendering the producer resistant to its own product. CalC resembles ATP-dependent fatty acid-CoA ligase FadDs, and it contains an N-terminal domain homologous to the LuxE superfamily, which is an ATP-dependent acyl protein synthase and catalyzes the esterification of fatty acids into metabolically active CoA thioesters (4). CalD resembles NAD(P)H-dependent oxidoreductases/dehydrogenases. CalF resembles 3-carboxymuconate cycloisomerases (21). CalH resembles chorismate mutases. The genes calU1 to calU5 had no strong similarity to genes of known functions in the database, and they could not be assigned any function in the biosynthesis of calcimycin.
DISCUSSION
Three gene replacement experiments and the predicted functions of 13 putative proteins, including 6 multifunctional PKS modules, indicate that we have cloned and sequenced the gene cluster coding for the biosynthesis of the very important divalent ionophore calcimycin.
Genes required for the biosynthesis of the pyrrole, spiroketal polyketide, and benzoxazole moieties (Fig. 1) have been clearly identified by the high similarities of the predicted proteins to known enzymes from other antibiotic biosynthesis pathways, and their functions are consistent with the results of precursor feeding experiments (39).
Four peripheral genes (calC, calD, calF, calH) have extensive end-to-end similarities to enzymes of known functions, but it is not certain that they are involved in calcimycin biosynthesis. The genes cal1 and cal29 are very similar to peptidases S15 and M20, respectively. Both genes are probably irrelevant to calcimycin production. It therefore seems likely that all the calcimycin biosynthetic genes are within the 64-kb gene cluster shown in Fig. 3 and almost certainly within the 100 kb spanned by the three cosmid clones.
Polyketide synthesis.
The five PKS genes of the cluster are not contiguous, and they are not arranged in the order of the biosynthetic steps. The program NRPS-PKS predicted that all the domains of CalA3 are deleted or inactive, and its role in calcimycin production remains unclear (2, 36). The remaining six PKS modules have five different predicted functions, giving unambiguous assignment functions to the steps of polyketide synthesis (Table 4). The predicted functions of the PKS modules thus fit exactly the chemical structure of calcimycin. This includes the selection of methylmalonate or malonate by the ACP modules, the redox state of each extender module, and the stereospecificity of the two chiral β-carbon groups, which is determined by the KR modules (Table 4). Both CalA2 and CalA5b (b indicates the second module of the PKS) were predicted to incorporate fully reduced propionate units into the growing polyketide chain (Table 4). Assuming that Cal5b was likely to be used after Cal5a, we tentatively named Cal5a PKS module 4, and Cal5b became PKS module 5 (Table 4). After 6 rounds of decarboxylative condensation using two malonyl-CoA moieties and four methylmalonyl-CoA moieties as extender units, the full-length polyketide chain must be detached from the final module of the assembly line.
A variety of termination strategies for PKS assembly lines are known (9). Most often, a C-terminal PKS TE domain catalyzes the release of the mature polyketide chain. Alternatively, the polyketide chain can be released by a separate type II TE, as exemplified by NanE for nanchangmycin, MonCII for monensin, and NigII for nigericin biosynthesis (9). The polyketide chain can also be released by a reductase domain or other discrete enzymes, such as pyridoxal-phosphate (PLP)-dependent 2-oxoamine synthase, actyltransferase, lactamase, or Baeyer-Villiger monooxygenase (9).
There is no TE domain at the C-terminal end of CalA4. Instead, we found CalG, a type II TE in the cal gene cluster. CalG may cleave the mature linear polyketide from CalA4, or, more efficiently, it may catalyze directly the intermolecular amide linkage between the amino group of 3-hydroxyanthranilic acid and the polyketide chain (Fig. 8, structure 3).
Synthesis of benzoxazole moiety.
Precursor feeding experiments showed that 3-hydroxyanthranilic acid is incorporated into the benzoxazole moiety (39). CalB1 to CalB4 are responsible for the biosynthesis of 3-hydroxyanthranilic acid. The extensive similarities of CalB4, CalB1, and CalB2 and PhzC, PhzE, and PhzD, respectively, suggest that this part of the calcimycin pathway is similar to the phenazine biosynthesis pathway (22). The precursor of benzoxazole for calcimycin might divert from 2,3-dihydro-3-hydroxyanthranilic acid, as shown in Fig. 7C. Disruption of calB2 totally abolished the production of calcimycin without obvious intermediate accumulation, providing no further clues for its function in precursor production (Fig. 5 and 6).
Synthesis of pyrrole moiety.
CalN1 to CalN3 were similar to CloN3 to CloN5, respectively, which are responsible for the biosynthesis of the pyrrole moiety from proline, as suggested by the results of feeding experiments (24, 39). Gene replacement and trans complementation confirmed the requirement of CalN genes for calcimycin biosynthesis.
Formation of spiroketal ring.
Spiroketal rings are found in tautomycin, avermectins, and spirandienes (10, 14, 18). It have been postulated that the spiroketal ring formation could proceed spontaneously by dual nucleophilic attack on the carbonyl carbon atom (C-14) by the two hydroxyl groups at C-10 and C-18 (Fig. 8, structure 6).
Tailoring steps.
Subsequent tailoring steps include hydroxylation, amination at C-3, and N-methylation of the benzoxazole moiety. N-methylation tailoring is catalyzed by CalM (Q. Wu, submitted). Removal of calM resulted in the accumulation of the calcimycin precursor (Fig. 8, structure 8). This proved that CalM was involved in post-PKS modification of cezomycin.
Regulation and resistance.
CalR1 and CalR2 resemble TylR-type and LuxR-type transcription regulators, respectively. Their location within the calcimycin gene cluster suggests that they participate in the regulation of calcimycin biosynthesis. The divergently transcribed genes calT and calR probably encode a calcimycin export pump and its regulator. They may also make the producer strain resistant to calcimycin.
Conclusions.
Calcimycin is special because of its biological activity and because it features a rare benzoxazole moiety. The identification and cloning of the cal biosynthetic gene cluster of S. chartreusis NRRL 3882 make it possible to plan the generation of calcimycin derivatives by genetic engineering involving gene replacements and expression of the entire gene cluster in different hosts.
Acknowledgments
We are grateful to Jim Swezey (ARS Patent Culture Collection) for kindly providing S. chartreusis NRRL 3882 and to Tobias Kieser for editing the manuscript.
We thank the National Science Foundation of China, the Ministry of Science and Technology (973 and 863 programs), the Ministry of Education of China, the Shanghai Municipal Council of Science and Technology and the Shanghai Leading Academic Discipline Project, the State Key Laboratory of Bio-organic and Natural Products Chemistry (CAS), and the National Program of Development of Transgenic New Species of China for research support.
Footnotes
Published ahead of print on 20 December 2010.
REFERENCES
- 1.Abbott, B. J., et al. 1979. Microbial transformation of A23187, a divalent cation ionophore antibiotic. Antimicrob. Agents Chemother. 16:808-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ansari, M. Z., G. Yadav, R. S. Gokhale, and D. Mohanty. 2004. NRPS-PKS: a knowledge-based resource for analysis of NRPS/PKS megasynthases. Nucleic Acids Res. 32:W405-W413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bierman, M., et al. 1992. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116:43-49. [DOI] [PubMed] [Google Scholar]
- 4.Black, P. N., C. C. DiRusso, A. K. Metzger, and T. L. Heimert. 1992. Cloning, sequencing, and expression of the fadD gene of Escherichia coli encoding acyl coenzyme A synthetase. J. Biol. Chem. 267:25513-25520. [PubMed] [Google Scholar]
- 5.Cerdeno, A. M., M. J. Bibb, and G. L. Challis. 2001. Analysis of the prodiginine biosynthesis gene cluster of Streptomyces coelicolor A3(2): new mechanisms for chain initiation and termination in modular multienzymes. Chem. Biol. 8:817-829. [DOI] [PubMed] [Google Scholar]
- 6.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.David, L., and S. Emadzadeh. 1982. Biosynthesis of the ionophorous antibiotic A23187. J. Antibiot. (Tokyo) 35:1616-1617. [DOI] [PubMed] [Google Scholar]
- 8.David, L., and A. Kergomard. 1982. Production by controlled biosynthesis of a novel ionophore antibiotic, cezomycin (demethylamino A23187). J. Antibiot. (Tokyo) 35:1409-1411. [DOI] [PubMed] [Google Scholar]
- 9.Du, L., and L. Lou. 2010. PKS and NRPS release mechanisms. Nat. Prod. Rep. 27:255-278. [DOI] [PubMed] [Google Scholar]
- 10.Frank, B., et al. 2007. Spiroketal polyketide formation in Sorangium: identification and analysis of the biosynthetic gene cluster for the highly cytotoxic spirangienes. Chem. Biol. 14:221-233. [DOI] [PubMed] [Google Scholar]
- 11.Garneau, S., P. C. Dorrestein, N. L. Kelleher, and C. T. Walsh. 2005. Characterization of the formation of the pyrrole moiety during clorobiocin and coumermycin A1 biosynthesis. Biochemistry 44:2770-2780. [DOI] [PubMed] [Google Scholar]
- 12.Gust, B., G. L. Challis, K. Fowler, T. Kieser, and K. F. Chater. 2003. PCR-targeting system in Streptomyces coelicolor. John Innes Center, Norwich, United Kingdom.
- 13.Heathcote, M. L., J. Staunton, and P. F. Leadlay. 2001. Role of type II thioesterases: evidence for removal of short acyl chains produced by aberrant decarboxylation of chain extender units. Chem. Biol. 8:207-220. [DOI] [PubMed] [Google Scholar]
- 14.Ikeda, H., T. Nonomiya, M. Usami, T. Ohta, and S. Omura. 1999. Organization of the biosynthetic gene cluster for the polyketide anthelmintic macrolide avermectin in Streptomyces avermitilis. Proc. Natl. Acad. Sci. U. S. A. 96:9509-9514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom.
- 16.Kim, B. S., T. A. Cropp, B. J. Beck, D. H. Sherman, and K. A. Reynolds. 2002. Biochemical evidence for an editing role of thioesterase II in the biosynthesis of the polyketide pikromycin. J. Biol. Chem. 277:48028-48034. [DOI] [PubMed] [Google Scholar]
- 17.Kozian, D., et al. 2005. Identification of genes involved in Ca2+ ionophore A23187-mediated apoptosis and demonstration of a high susceptibility for transcriptional repression of cell cycle genes in B lymphoblasts from a patient with Scott syndrome. BMC Genomics 6:146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li, W., J. Ju, S. R. Rajski, H. Osada, and B. Shen. 2008. Characterization of the tautomycin biosynthetic gene cluster from Streptomyces spiroverticillatus unveiling new insights into dialkylmaleic anhydride and polyketide biosynthesis. J. Biol. Chem. 283:28607-28617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu, C. M., et al. 1983. X-14885A, a novel divalent cation ionophore produced by a Streptomyces culture: discovery, fermentation, biological as well as ionophore properties and taxonomy of the producing culture. J. Antibiot. (Tokyo) 36:1118-1122. [DOI] [PubMed] [Google Scholar]
- 20.Liu, C. M., et al. 1979. X-14547A, a new ionophorous antibiotic produced by Streptomyces antibioticus NRRL 8167. Discovery, fermentation, biological properties and taxonomy of the producing culture. J. Antibiot. (Tokyo) 32:95-99. [DOI] [PubMed] [Google Scholar]
- 21.Lorite, M. J., J. Sanjuan, L. Velasco, J. Olivares, and E. J. Bedmar. 1998. Characterization of Bradyrhizobium japonicum pcaBDC genes involved in 4-hydroxybenzoate degradation. Biochim. Biophys. Acta 1397:257-261. [DOI] [PubMed] [Google Scholar]
- 22.Mavrodi, D. V., et al. 1998. A seven-gene locus for synthesis of phenazine-1-carboxylic acid by Pseudomonas fluorescens 2-79. J. Bacteriol. 180:2541-2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nowak-Thompson, B., N. Chaney, J. S. Wing, S. J. Gould, and J. E. Loper. 1999. Characterization of the pyoluteorin biosynthetic gene cluster of Pseudomonas fluorescens Pf-5. J. Bacteriol. 181:2166-2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pojer, F., S. M. Li, and L. Heide. 2002. Molecular cloning and sequence analysis of the clorobiocin biosynthetic gene cluster: new insights into the biosynthesis of aminocoumarin antibiotics. Microbiology 148:3901-3911. [DOI] [PubMed] [Google Scholar]
- 25.Pressman, B. C. 1976. Biological applications of ionophores. Annu. Rev. Biochem. 45:501-530. [DOI] [PubMed] [Google Scholar]
- 26.Prudhomme, M., G. Dauphin, J. Guyot, and G. Jeminet. 1984. Semisynthesis of A23187 (calcimycin) analogs. II. Introduction of a methyl group on the benzoxazole ring. J. Antibiot. (Tokyo) 37:627-634. [DOI] [PubMed] [Google Scholar]
- 27.Prudhomme, M., G. Dauphin, and G. Jeminet. 1986. Semi-synthesis of A23187 (calcimycin) analogs. III. Modification of benzoxazole ring substituents, ionophorous properties in an organic phase. J. Antibiot. (Tokyo) 39:922-933. [DOI] [PubMed] [Google Scholar]
- 28.Prudhomme, M., J. Guyot, and G. Jeminet. 1986. Semi-synthesis of A23187 (calcimycin) analogs. IV. Cation carrier properties in mitochondria of analogs with modified benzoxazole rings. Antimicrobial activity. J. Antibiot. (Tokyo) 39:934-937. [DOI] [PubMed] [Google Scholar]
- 29.Reed, P. W., and H. A. Lardy. 1972. A23187: a divalent cation ionophore. J. Biol. Chem. 247:6970-6977. [PubMed] [Google Scholar]
- 30.Reid, R., et al. 2003. A model of structure and catalysis for ketoreductase domains in modular polyketide synthases. Biochemistry 42:72-79. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 32.Schubert, H. L., R. M. Blumenthal, and X. Cheng. 2003. Many paths to methyltransfer: a chronicle of convergence. Trends Biochem. Sci. 28:329-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thomas, M. G., M. D. Burkart, and C. T. Walsh. 2002. Conversion of l-proline to pyrrolyl-2-carboxyl-S-PCP during undecylprodigiosin and pyoluteorin biosynthesis. Chem. Biol. 9:171-184. [DOI] [PubMed] [Google Scholar]
- 34.Wang, Z. X., S. M. Li, and L. Heide. 2000. Identification of the coumermycin A(1) biosynthetic gene cluster of Streptomyces rishiriensis DSM 40489. Antimicrob. Agents Chemother. 44:3040-3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Westley, J. W., et al. 1983. Isolation and characterization of a novel polyether antibiotic of the pyrrolether class, antibiotic X-14885A. J. Antibiot. (Tokyo) 36:1275-1278. [DOI] [PubMed] [Google Scholar]
- 36.Yadav, G., R. S. Gokhale, and D. Mohanty. 2003. Computational approach for prediction of domain organization and substrate specificity of modular polyketide synthases. J. Mol. Biol. 328:335-363. [DOI] [PubMed] [Google Scholar]
- 37.Zhang, X., and R. J. Parry. 2007. Cloning and characterization of the pyrrolomycin biosynthetic gene clusters from Actinosporangium vitaminophilum ATCC 31673 and Streptomyces sp. strain UC 11065. Antimicrob. Agents Chemother. 51:946-957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou, Y., et al. 2008. Selective removal of aberrant extender units by a type II thioesterase for efficient FR-008/candicidin biosynthesis in Streptomyces sp. strain FR-008. Appl. Environ. Microbiol. 74:7235-7242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zmijewski, M. J., Jr. 1980. Biosynthesis of antibiotic A23187. Incorporation of precursors into A23187. J. Antibiot. (Tokyo) 33:447-450. [DOI] [PubMed] [Google Scholar]