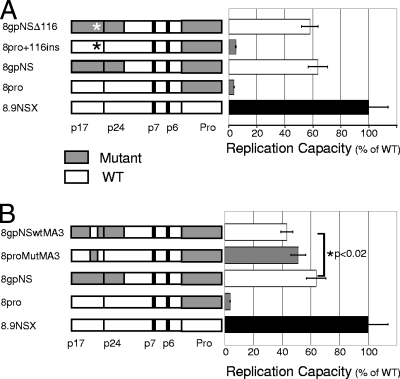
FIG. 1.
Replication capacity of mutant matrix and partial capsid constructs with mutant protease. Recombinant resistance vectors containing wild-type (white) and mutant (shaded) regions were titrated by serial dilution. The luciferase signal is shown relative to wild type after normalization for p24 protein levels. The names and schematic representations of gag-protease constructs are shown. Protease cleavage sites are shown: cleavage sites and p1 and p2 spacer peptides are depicted by heavy lines. The functional Gag proteins matrix (p17), capsid (p24), nucleocapsid (p7), and p6 (p1 and p2 not shown) are listed, as is protease (Pro). Error bars show the standard error of the mean. The mean was derived from 11 data points for all constructs, except 8pro, which was derived from 9 data points. (A) Effect of mutant insertion at amino acid 116 of matrix, shown by an asterisk in the schematic (8pro+116ins and 8gpNSΔ116). (B) The effect of changing three amino acids together at positions 76, 79, and 81 of matrix is shown as a gray box (mutant) in an otherwise white wild-type gag (8proMutMA3) and as a white box (WT) in an otherwise gray mutant matrix (8gpNSwtMA3).