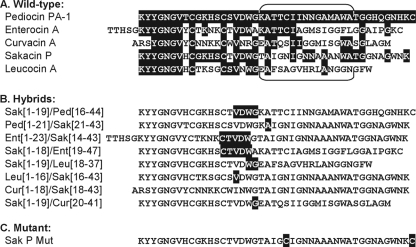
FIG. 1.
(A) Amino acid sequences of the wild-type pediocin-like bacteriocins used in this study. Regions where the sequences are identical to the sequence in pediocin PA-1 are in white with a black background. The brackets above and below the sequences indicate the regions from which the 15-mer fragments were derived. The Ped-15-mer, Sak-15-mer, and Leu-15-mer fragments span residues 20 to 34 of pediocin PA-1, sakacin P, and leucocin A, respectively, whereas the Ent-15-mer fragment spans residues 25 to 34 of enterocin A and the Cur-15-mer fragment spans residues 21 to 35 of curvacin A. (B) Amino acid sequences of the hybrid bacteriocins used in this study. The Sak[1-19]/Ped[16-44] hybrid consists of the N-terminal half of sakacin P (residues 1 to 19) and the C-terminal half of pediocin PA-1 (residues 16 to 44). Residues 16 to 19 of sakacin P and pediocin PA-1 are identical and are in white with a black background. The terminology and use of white with a black background are analogous for the other hybrid bacteriocins. Note that there is a conservative mutation, V17I, in the Cur[1-18]/Sak[18-43] hybrid. (C) Amino acid sequences of the sakacin P variant. This variant has a disulfide bridge in its C-terminal half, since two cysteine residues have been introduced into sakacin P, at positions 24 and 44, both indicated in white with a black background. (The sequences are based on data from references 18, 23, and 24 for pediocin PA-1; reference 3 for enterocin A; references 19 and 31 for curvacin A; reference 30 for sakacin P; and reference 15 for leucocin A.)