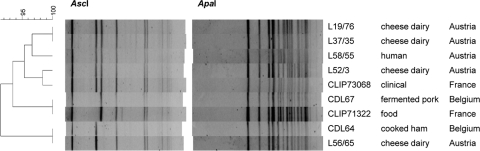
FIG. 2.
PFGE analysis of L. monocytogenes strains harboring lin0464 and lin0465 homologues. Cluster analysis was performed with Fingerprinting II software (Bio-Rad, Hercules, CA) using the following settings: 0.2% optimization, 0.5% position tolerance, and the Dice similarity coefficient. For dendrogram construction, the UPGMA (unweighted-pair group method using average linkages) algorithm was used. Strain designations, sources, and countries of origin are given on the right, and percentages of similarity are indicated on the left.