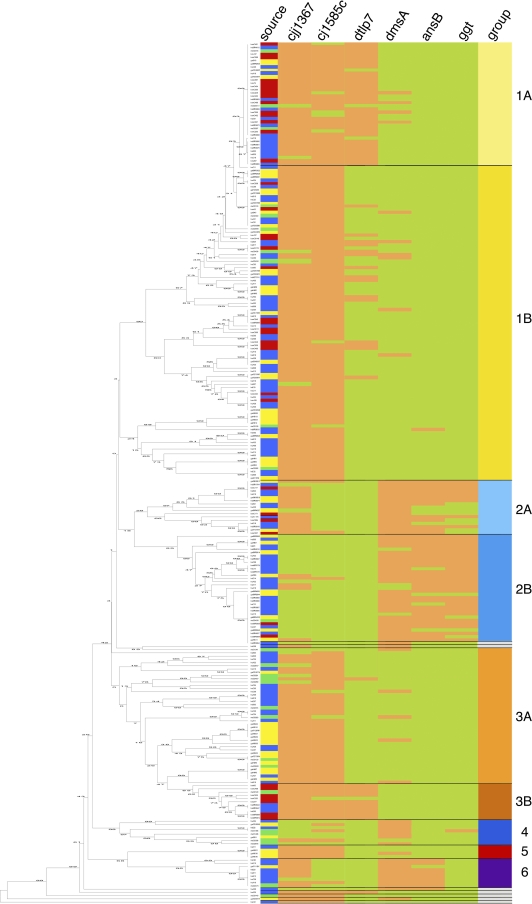
FIG. 3.
MLST-based UPGMA tree and arrangement of the six different marker genes within the six defined groups (nine subgroups). The MLST-based UPGMA tree for 266 C. jejuni isolates is depicted on the left. The numbers shown on the branches of the tree indicate the linkage distances. The right side shows a table of all isolates in the order of the UPGMA tree depicting the source of the isolate, the presence or absence of the six marker genes, and their association with one of the groups defined in Table 1. Sources: blue, human isolates; yellow, chicken isolates; red, bovine isolates; green, turkey isolates. The presence of a genetic marker is marked with light red, and its absence is marked with light green. The genetic markers, from left to right, are as follows: cjj1367, cjj81176-1367/1371 gene (cj1365c), encoding a serine protease; cj1585c, gene encoding an oxidoreductase and replacing dmsA to -D in NCTC 11168; dtlp7, cj0951c and cj0952c genes, encoding the heterodimeric form of transducer-like protein 7; dmsA, gene encoding the dimethyl sulfoxide oxidoreductase subunit A; ansB, gene encoding an asparaginase with an accessory N-terminal sec-dependent secretion signal for periplasmic localization of the enzyme; ggt, gene encoding the γ-glutamyl-transpeptidase. Groups: light yellow, 1a; intense yellow, 1b; cyan blue, 2a; bondi blue, 2b; carrot orange, 3a; orange-red, 3b; blue, 4; red, 5; purple, 6; white, singletons.