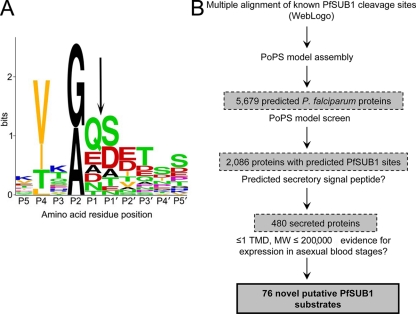
FIG. 1.
Predicted substrate preference of PfSUB1 and its application to an in silico search for new substrates. (A) Graphical representation in single-letter code of a multiple-sequence alignment of amino acid residues flanking known and predicted PfSUB1 cleavage sites within SERA5 (sites 1, 2, and 3), SERA4 and SERA6 (predicted sites 1 and 2), and MSP1, MSP6, and MSP7 (a total of 17 different sequences), plus the internal PfSUB1 autocatalytic processing site at which cleavage occurs during protease maturation (66). The overall height of each stack of residues indicates the degree of sequence conservation at that position, while the height of each residue within the stack indicates the relative frequency of each amino acid residue at that position. Residues are color coded according to the chemical nature of their side chains (red, acidic [D and E]; blue, basic [K, R, and H]; orange, aliphatic [L, V, and I]; black, small [G and A]; green, uncharged polar [S, T, Y, N, and Q]; and purple, nonpolar, nonaliphatic [F and P]). The scissile bond is indicated by an arrow. Residue numbering is according to the system of Schechter and Berger (68). The figure was produced using the WebLogo facility at http://weblogo.berkeley.edu/logo.cgi and annotated with Adobe Photoshop. (B) Overview of PoPS-based in silico screen for candidate new PfSUB1 substrates. TMD, transmembrane domain.