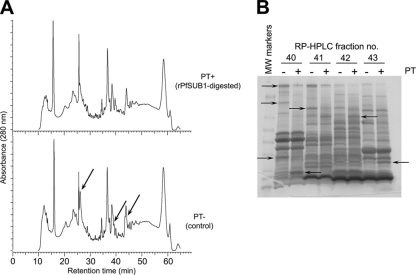
FIG. 3.
Fractionation of rPfSUB1-digested schizont extracts reveals protein substrates of PfSUB1. (A) Comparative C4 RP-HPLC elution profiles of rPfSUB1-digested (PT+) and control (PT-) schizont membrane extracts. The profiles were largely very similar, consistent with cleavage of only a small fraction of the total protein population by rPfSUB1 (see Fig. S1A in the supplemental material). However, some differences were evident (arrows). Similar results were obtained upon RP-HPLC fractionation of ST+ and ST− samples (data not shown). (B) Representative InstantBlue-stained SDS-PAGE gel of RP-HPLC-fractionated PT+ and PT− samples, indicating species modified by incubation with rPfSUB1 (arrows) as determined by visual comparison of equivalent RP-HPLC eluate fractions. The complete set of SDS-PAGE-analyzed RP-HPLC fractions, annotated with protein identities as determined by LC-MS-MS of excised bands, is shown in Fig. S2 in the supplemental material. MW, molecular weight.