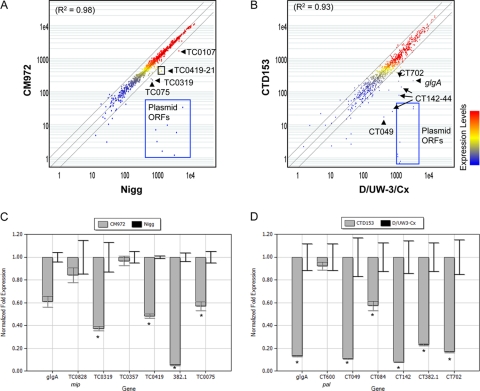
FIG. 4.
Transcriptional profiling via microarray screening and quantitative RT-PCR of plasmid-deficient C. muridarum and C. trachomatis reveals a subpopulation of plasmid-responsive genes. (A and B) Scatter plot illustrations of microarray comparisons of C. muridarum Nigg with strain CM972 (A) and C. trachomatis D/UW3-Cx with CTD153 (B). (C and D) RNA was isolated from infected cells 30 h after infection. Quantitative RT-PCR confirmed reduced transcription of candidate PRCL in C. muridarum CM972 (C) and C. trachomatis CTD153 (D). Total RNA was isolated from infected cells 24 h after infection, and PRCL transcripts were measured by quantitative RT-PCR. The transcriptional differences are presented as the fold change in expression for each gene in a single representative experiment, although each experiment was performed independently at least twice and samples were assayed in triplicate. *, P ≤ 0.013 (Nigg versus CM972) or P ≤ 0.025 (D/UW-3Cx versus CTD153). TC0828 (mip) or CT600 (pal) are included as examples of non-plasmid-responsive controls, because we did not detect a difference in the transcription of these genes by microarray screen.