Abstract
Cronobacter spp. are emerging neonatal pathogens in humans, associated with outbreaks of meningitis and sepsis. To cause disease, they must survive in blood and invade the central nervous system by penetrating the blood-brain barrier. C. sakazakii BAA-894 possesses an ∼131-kb plasmid (pESA3) that encodes an outer membrane protease (Cpa) that has significant identity to proteins that belong to the Pla subfamily of omptins. Members of this subfamily of proteins degrade a number of serum proteins, including circulating complement, providing protection from the complement-dependent serum killing. Moreover, proteins of the Pla subfamily can cause uncontrolled plasmin activity by converting plasminogen to plasmin and inactivating the plasmin inhibitor α2-antiplasmin (α2-AP). These reactions enhance the spread and invasion of bacteria in the host. In this study, we found that an isogenic cpa mutant showed reduced resistance to serum in comparison to its parent C. sakazakii BAA-894 strain. Overexpression of Cpa in C. sakazakii or Escherichia coli DH5α showed that Cpa proteolytically cleaved complement components C3, C3a, and C4b. Furthermore, a strain of C. sakazakii overexpressing Cpa caused a rapid activation of plasminogen and inactivation of α2-AP. These results strongly suggest that Cpa may be an important virulence factor involved in serum resistance, as well as in the spread and invasion of C. sakazakii.
Cronobacter spp., formerly known as Enterobacter sakazakii, are Gram-negative rod-shaped bacteria within the family Enterobacteriaceae. The genus Cronobacter comprises five species—C. sakazakii, C. malonaticus, C. turicensis, C. muytjensii, and C. dublinensis—and genomospecies group 1. They are emerging pathogens that cause severe meningitis, septicemia, or necrotizing enterocolitis in neonates and infants (22, 38, 61). Although the disease frequency is very low, the mortality rate ranges from 40% to as high as 80% (12, 38, 61). Meningitis caused by Cronobacter spp. occurs both as sporadic cases and as outbreaks, and contaminated powdered infant formulas have been epidemiologically implicated as the source of the pathogen in most cases (2, 16, 32, 51). In order to cause meningitis, it is expected that Cronobacter spp. express virulence factors that help in colonization of the mucosal surfaces, allow for the translocation into the bloodstream and overcome host defense mechanisms.
Little is known about the virulence factors of Cronobacter spp. and the pathogenic mechanisms involved in neonatal infections. Using suckling mice, it was demonstrated that Cronobacter spp. produce an enterotoxin when orally fed; however, the genes encoding the putative toxin have yet to be identified (39, 42). Kothary et al. (23) found that all Cronobacter spp. produce a zinc-containing metalloprotease that causes rounding of CHO cells. It has also been shown that the outer membrane protein A (OmpA) of Cronobacter spp. is involved in the colonization of the gastrointestinal tract and invasion of human intestinal epithelial and brain endothelial cells, as well as subsequent survival in blood to cause meningitis (36, 37, 53). Recently, it was reported that the outer membrane proteins OmpX and OmpA are involved in the basolateral invasion of enterocyte-like human epithelial cells by C. sakazakii (21).
Whole-genome sequencing of C. sakazakii BAA-894 showed that this strain possesses two plasmids, pESA2 and pESA3 (24). By using in silico analysis, we determined that pESA3 encodes an outer membrane protease with significant identity to proteins that belong to the omptin family. The omptins include a family of aspartate proteases that are surface-orientated outer membrane proteins expressed by many members of the Enterobacteriaceae (26, 18). Most of the known omptins are bacterial virulence factors and function as proteases, adhesins, or invasins (14, 18, 26).
Amino acid sequence analyses of known omptin proteins predict that there are two groups within the family. The first group, called the Pla subfamily, consists of the PgtE of Salmonella enterica, Pla of Yersinia pestis, and PlaA of Erwinia spp. The second subfamily, designated OmpT, contains OmpT and OmpP of Escherichia coli and SopA of Shigella flexneri (14, 26). Moreover, studies have shown that the members in each subfamily are functionally similar (14, 26). PgtE and Pla are involved in uncontrolled plasmin activity by efficient conversion of human proenzyme, plasminogen to plasmin, inactivation of the plasmin inhibitors α2 antiplasmin (α2-AP) and plasminogen activator inhibitor 1 (PAI-1) (13, 27, 29, 56). These properties enhance the spread and multiplication of Y. pestis and S. enterica in the host (26, 27, 28, 49, 55). Pla and PgtE also degrade a number of serum proteins, including circulating complement, providing Y. pestis and S. enterica protection from complement-dependent serum killing (44, 55). PlaA is likely to play an important role in bacterium-plant interactions. Members of the OmpT subfamily, including OmpT and SopA, slowly cleave plasminogen (33, 34) to plasmin, which is then quickly inhibited by α2-AP (27). OmpT and SopA have also been found to play a role in bacterial virulence in ways that are unrelated to plasminogen activation. OmpT of E. coli causes the cleavage of protamine and other cationic peptides that possess antibiotic activity (11, 57), and SopA of S. flexneri cleaves IcsA, which is an important requirement in the intercellular spread of shigellae into adjacent host cells by means of actin-based motility (5, 50).
Alignment of the amino acid sequence of the C. sakazakii outer membrane protease with different members of the omptin family indicates that the protease belongs to the Pla subfamily. Due to the high similarity with Pla, we named this outer membrane protease “Cpa,” which stands for “Cronobacter plasminogen activator.” In the present study, we investigated the role of Cpa in providing protection to C. sakazakii from complement-dependent serum killing, as well as in activating plasminogen and inactivating α2-AP.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains and plasmids used in the present study are described in Table 1 . A strain of C. sakazakii BAA-894, which was resistant to nalidixic acid (BAA894NA) was isolated as a spontaneous mutant using the method described by Johnson et al. (20). Cronobacter spp. and E. coli strains were grown at 37°C in Luria-Bertani (LB) broth with shaking (175 rpm) or on LB agar. Antibiotics were added when required at the following concentrations: ampicillin (100 μg/ml) and nalidixic acid (256 μg/ml).
TABLE 1.
Bacteria and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s)a | Source or reference |
|---|---|---|
| Strains | ||
| C. sakazakii | ||
| BAA-894 | Isolated from powdered infant formula associated with a NICU outbreak, obtained from the ATCC (Manassas, VA) | 16 |
| BAA894NA | BAA-894 nalidixic acid-resistant spontaneous mutant | This study |
| BAA894Δcpa | BAA-894 cpa isogenic mutant | This study |
| BAA894Δcpa/pMMB66EH::cpa | BAA894Δcpa complemented with pMMB66EH::cpA | This study |
| E. coli | ||
| BL21 | F−dcm ompT hsdS(rB− mB−) gal [malB+]K-12(λS) | Novagen |
| DH5α | F− φ80lacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17 (rK− mK+) phoA supE44 λ−thi-1 gyrA96 relA1 | Invitrogen |
| DH5α/pMMB66EH | DH5α containing plasmid pMMB66EH | This study |
| DH5α/pMMB66EH::cpa | DH5α containing pMMB66EH::cpa | This study |
| DH5α λpir | DH5α containing λpir | 52 |
| SM10 λpir | thi thr leu tonA lacY supE recA::RP4-2-Tc::Mu Km λpir | 4 |
| Plasmids | ||
| pET30(+) | Expression vector, T7 promoter-driven system | Novagen |
| pET30::cpa | Plasmid expressing His6-CpA fusion protein | This study |
| pCVD442 | Suicide vector, R6K ori, mobRP4, bla, sacB | 4 |
| pMMB66EH | Broad-host-range Ptac expression vector | 10 |
| pMMB66EH::cpa | cpa cloned into pMMB66EH | This study |
| pCVD442::Δcpa | Δcpa and flanking region cloned into pCVD442 | This study |
NICU, neonatal intensive care units; ATCC, American Type Culture Collection.
Construction of cpa isogenic mutant.
Nucleotide sequence of plasmid pESA3 was obtained from the GenBank database (http://www.ncbi.nlm.nih.gov/GenBank/index.html; accession no. NC_009780). The sequence of the gene ESA_pESA3p05434 (named cpa in the present study) and flanking regions were used to design primers for isolation of a cpa mutant and cloning of cpa (24). A cpa isogenic mutant was created in C. sakazakii BAA-894 by using the method previously described for the mutation of the Bacteroides fragilis toxin gene (46). Briefly, a primer internal to and orientated upstream of cpa (primer mut2; XhoI, 5′-GCGGATGACTCGAGTGTTACAGAAGAAGCGGCATTCGC) was used in a PCR with primer mut1 (XbaI, 5′-CGACGGACTCTAGACTGGAGTGTGGACTGGGCGCTTTATG) located ∼3 kb upstream of primer mut2 (the restriction sites are underlined). A second PCR used a primer within cpa orientated downstream (primer mut3; XhoI, 5′-GCGCGACTCTCGAGATGATTAATAACGCGACCGGAACGTC) with a primer located ∼3 kb downstream of primer 3 (primer mut4; XbaI, 5′-GCGCCACTTCTAGATCTGGTGCGCACCTTTGATGCGCTGC; the restriction sites are underlined). PCRs with primers mut1 and mut2 and primers mut3 and mut4 were performed with AccuPrime Taq DNA Polymerase High Fidelity (Invitrogen). The PCR products were digested with XbaI and XhoI and cloned by three-way ligation into the suicide vector pCVD442 at the XbaI site (4). Ligation of the XhoI sites created an in-frame deletion, removing 87% of the 936 bp of cpa. E. coli DH5α λpir was transformed with the ligated DNA sample and a plasmid containing the PCR products in the correct orientation was selected (pCVD442::Δcpa) by PCR and sequenced to demonstrate mutation of cpa. Then pCVD442::Δcpa was transformed into E. coli SM10 λpir and then mobilized into C. sakazakii BAA894NA. Single homologous recombination clones were selected in LB-agar containing ampicillin and nalidixic acid and, subsequently, double homologous recombination mutants were selected in LB-agar containing 10% sucrose as described by Donnenberg and Kaper (4). Δcpa clones, which were sucrose resistant and ampicillin sensitive, were confirmed by PCR, sequence analyses, and Western blot analysis with rabbit anti-His6-Cpa antiserum as described below.
Complementation.
For complementation of cpa mutant, the cpa gene was amplified by PCR using the primers Cpafw (EcoRI, 5′-GCCTGGCGGAATTCAATGGAATAATATATGAATAAGAAACTTATTGTCG) and Cparv (PstI, 5′-GATCAAAGCTGCAGTCAGAAACGGTACTGAAGACCTGCGG), digested with EcoRI and PstI, and cloned into the Ptac expression vector pMMB66EH (10). The resultant plasmid pMMB66EH::cpa was transformed into E. coli SM10 λpir and then mobilized into BAA894Δcpa by conjugation. Expression of Cpa by pMMB66EH::cpa using BAA894Δcpa or DH5α as hosts was induced by 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) and confirmed by Western blot analysis with rabbit anti-His6-Cpa antiserum as described below.
Cpa purification and production of anti-Cpa antiserum.
In order to produce antibodies against CpA, a His6-Cpa fusion protein was cloned and expressed in E. coli. Briefly, cpa was cloned into pET30(+) T7 promoter-based expression vector (Novagen, San Diego, CA) by PCR amplification by using the primers EcoRI, (5′-GCCTGGCGGAATTCATGAATAAGAAACTTATTGTCGTGGCGA) and XhoI (5′-CGGCTCGCCTCGAGTCAGAAACGGTACTGAAGACCTGCGG). The recombinant plasmid was transformed into E. coli BL21, and the overexpressed His6-Cpa fusion protein was purified under denaturing conditions by chromatography using a Ni-NTA kit according the manufacturer's instructions (Qiagen, Valencia, CA). Antibodies against His6-Cpa protein were prepared by injecting 1 mg of fusion proteins emulsified with RIBI adjuvant system (Corixa, Hamilton, MT) into rabbits according to FDA/IACUC protocol number 013X. A booster of 500 μg of fusion protein and adjuvant was injected 4 weeks after the first injection. Anti-Cpa antibodies titers were measured by using a standard enzyme-linked immunosorbent assay procedure using protein A-peroxidase, hydrogen peroxide, and ABTS [2,2′azinobis(3-ethylbenzthiazolinesulfonic acid)].
Identification of Cpa.
C. sakazakii and E. coli cultures grown for18 h in LB broth were collected by centrifugation and suspended in phosphate-buffered saline (PBS; pH 7.4). Bacterial cells were lysed by sonication, and cell debris was pelleted by centrifugation. Whole-cell lysates (4 μg) were separated by SDS-PAGE using 8 to 25% gradient PhastGels (GE Healthcare, Piscataway, NJ) and electro blotted onto nitrocellulose membranes. Cpa expression was confirmed by using anti-His6-Cpa antisera (1:500 dilution) as the primary antibody, goat anti-rabbit IgG-alkaline phosphatase conjugate as the second antibody (1:3,000 dilution), and an alkaline phosphatase substrate detection kit (Bio-Rad, Hercules, CA).
Serum survival assay.
Wild-type and recombinant C. sakazakii and E. coli strains were grown to mid-logarithmic phase, collected by centrifugation, and washed and diluted in PBS (pH 7.4). C. sakazakii (106 CFU) and E. coli (107 CFU) cultures were suspended in 12 and 25% of normal human serum (NHS), followed by incubation at 37°C for 1 h. The bacteria were then serially diluted and plated on LB-agar for quantitative determination. Serum heated at 56°C for 30 min (heat-inactivated serum; HIS) was used as control in all experiments. To differentiate which potential complement activation pathway may be involved in serum resistance, NHS was incubated with 20 mM EGTA plus 5 mM MgCl2 for 30 min at 25°C for classical and lectin pathway inactivation (classical and lectin-dependent complement pathway-inactivated serum [CCPIS]) (3, 8) or heated at 47°C for 20 min for alternative pathway inactivation (alternative complement pathway inactivated serum [ACPIS]) (35). The percent survival was calculated as follows: (the average number of bacteria that survive exposure to NHS or partially inactivated serum at 1 h divided by the number of bacteria that survive exposure to HIS at 1 h) × 100.
Proteolysis of complement proteins C3, C3a, and C4b.
Portions (2 ml) of wild-type and recombinant C. sakazakii and E. coli cultures grown to mid-logarithmic phase were collected by centrifugation and resuspended in 0.2 ml of PBS (pH 7.4). To determine proteolysis of purified C3, C3a, and C4b, 6 μl of the bacterial suspension (6 × 107 cells) was added to 4 μl of PBS (pH 7.4) containing 1.8 μg of purified human C3, C3a, or C4b (Calbiochem, San Diego, CA), followed by incubation for 18 h and 45 min at 37°C. To determine the proteolysis of C3 in NHS, 10 μl of bacterial suspension (108 cells) was mixed with 5 μl of PBS and 5 μl of NHS (final concentration of NHS, 25%), followed by incubation for 18 h and 30 min at 37°C. After incubation, the samples were centrifuged, and supernatants were separated by SDS-PAGE using either 12.5% homogeneous (to identify cleavage of C3) or 8 to 25% gradient (to identify cleavage of C3a and C4b) gels, electro blotted to nitrocellulose membrane, and probed with goat anti-human C3 or C4b (Calbiochem) antibodies. Alkaline phosphatase-conjugated rabbit anti-goat antibody was used as a secondary antibody to detect the cleaved complement proteins.
Plasminogen activation.
Kinetic measurement of plasminogen activation was performed by incubating 3 × 107 to 3 × 108 bacteria, 4 μg of human-Glu-plasminogen (Hematologic Technologies, Inc., River Road, VT), and the chromogenic plasmin substrate Val-Leu-Lys-p-nitroaniline dihydrochloride (0.5 mM S-2251; Chromogenix) in a total volume of 200 μl at 37°C. Plasmin formation was measured every 20 min at 405 nm by using a microtiter plate reader (Molecular Devices, Sunnyvale, CA). Plasmin (Sigma-Aldrich) was used as a positive control at a final concentration of 20 μg/ml. The effect of α2-AP on plasmin activity was assayed at a final concentration of 25 μg/ml.
Phylogenetic analysis.
Phylogenetic analyses of omptins nucleotide sequences were conducted in MEGA4 (60). The evolutionary history was inferred by using the neighbor-joining method (48). The bootstrap consensus tree inferred from 500 replicates (7) is taken to represent the evolutionary history of the taxa analyzed (7). The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (500 replicates) are shown next to the branches (7). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed by using the Poisson correction method (63) and are in the units of the number of amino acid substitutions per site. All positions containing gaps and missing data were eliminated from the data set. There were a total of 309 positions in the final data set.
N-terminal amino acid sequencing.
The ∼100-kDa protein band produced by proteolysis of the C3α was excised from a Western blotted Problott membrane and stained with Coomassie brilliant blue R250, and the N-terminal amino acid sequence was determined by Edmund degradation using a Procise model 492 protein Sequencer (Applied Biosystems, Foster City, CA).
Accession numbers.
The NCBI accession numbers of the nucleotide and proteins sequences used in the present study are as follows: Pla, NP_857784; PgtE, AF239770; PlaA, NP_857613; OmpT, AP_001210; OmpP, X74278; and SopA, NP_858404. The NCBI accession number of pESA3 is NC_009780, and cpa has the gene number ESA_pESA3p05434.
RESULTS
In silico sequence analysis of Cpa.
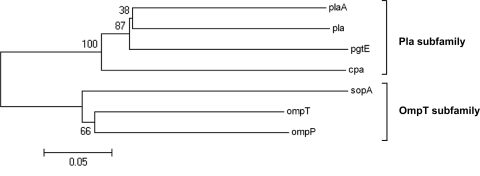
Phylogenetic cluster analysis of the nucleotide sequence of C. sakazakii cpa revealed that cpa clusters with the Pla subfamily of omptins (Fig. 1). The omptin proteases contain a conserved barrel structure with 10 transmembrane β-strands and five surface-exposed loops (L1 to L5) (26, 27, 62). Mutagenesis scanning, loop swapping, and substitution analyses showed that the differing polypeptide substrate selectivity of omptins is dictated by sequence variation in the surface-exposed loops of the β-barrel (15, 27, 43). Alignment of the amino acid sequence of Cpa with those of Pla, PgtE, and OmpT showed that the predicted Cpa surface-exposed loops share a higher identity with those of Pla and PgtE than with OmpT (Fig. 2). A recent study showed that the most variable surface residues between Pla and OmpT subfamilies are T36, L213, I260, D266, and D273, suggesting that these amino acids are important for substrate specificity (13; J. Haiko, unpublished data). Sequence alignment of Cpa with Pla and PgtE showed that all of these predicted substrate specific amino acids, except for D266, are conserved in Cpa (Fig. 2). The sequence similarity of Cpa with members of the Pla subfamily of omptins led to our hypothesis that Cpa may have similar functions to those of Pla and/or PgtE.
FIG. 1.
Phylogenetic tree of omptin family members. Phylogenetic analyses were conducted in MEGA4 (60). The evolutionary history was inferred using the neighbor-joining method (48).
FIG. 2.
Protein sequence alignment of C. sakazakii Cpa with known omptins: Pla of Y. pestis, OmpT of E. coli, and PgtE of S. enterica. Sequences were aligned by using the MEGA4 program (60). Residues important for proteolytic activity are in boldface, putative important residues for Pla subfamily substrate specificity are in marked by an asterisk (13), and the predicted surface-exposed loops determined for OmpT are indicated by L1 to L5 (62). Amino acid residues are numbered according to Pla sequence.
Generation of a C. sakazakii cpa deletion mutant.
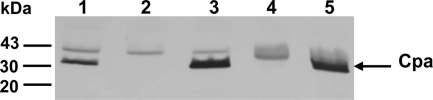
In order to determine whether Cpa is required for serum resistance and virulence of C. sakazakii, a cpa isogenic mutant, BAA894Δcpa, was constructed. To confirm that the deletion was in-frame deletion PCR with specific primers targeting internal and flanking regions of cpa was performed and the PCR product was sequenced (data not shown). Western blot analysis with antibodies against the His6-Cpa fusion protein revealed the presence of Cpa (∼33 kDa) in wild-type (WT) strain BAA-894, but not in mutant strain BAA894Δcpa (Fig. 3, lanes 1 and 2).
FIG. 3.
Immunoblot of Cpa. Similar amounts (4 μg) of whole-cell lysate preparations of C. sakazakii and E. coli strains were analyzed by Western blotting with a rabbit anti-His6-Cpa serum. Lanes: 1, BAA-894; 2, BAA894Δcpa; 3, BAA894Δcpa/pMMB66EH::cpa; 4, DH5α/pMMB66EH; 5, DH5α/pMMB66EH::cpa. Molecular masses (in kilodaltons) are indicated on the left.
Cpa is required for resistance to complement-mediated killing.
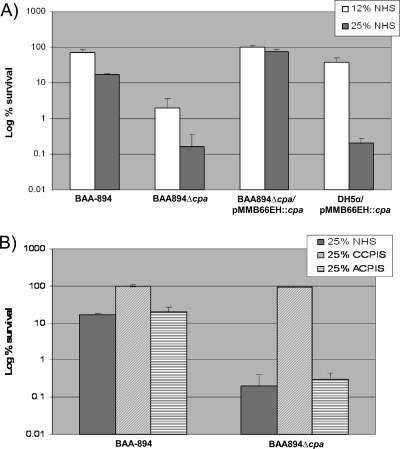
The C. sakazakii BAA894Δcpa mutant and WT BAA-894 were tested for survival in the presence of NHS and HIS. Figure 4A shows that after incubation for 1 h with 12% NHS, 2% of mutant BAA894Δcpa survived compared to the 70% survival rate of WT BAA-894. When the serum concentration was increased to 25%, the survival rate declined to 0.16% for BAA894Δcpa and to 17% for BAA-894 (Fig. 4A).
FIG. 4.
C. sakazakii Cpa is required for survival in NHS. (A) C. sakazakii and recombinant DH5α strains were incubated with 12 and 25% of NHS. (B) C. sakazakii BAA-894 and BAA894Δcpa strains were incubated with 25% CCPIS and 25% ACPIS. A serum survival assay was performed as described in Materials and Methods. The percent survivals shown are the means ± the standard deviations of three different assays. The P value was <0.05 for comparison of BAA-894 and BAA894Δcpa survival in 25 and 12% NHS as calculated by using the Student t test.
The cpa mutation was complemented by cloning the cpa gene into the expression vector pMMB66EH and mobilizing this construct into BAA994Δcpa. IPTG induction of Cpa expression in BAA994Δcpa/pMMB66EH::cpa resulted in 99 and 73% survival in the presence of 12 and 25% NHS, respectively (Fig. 4A). Western blot analysis with antibodies against the His6-Cpa fusion protein showed that the BAA994Δcpa/pMMB66EH::cpa complemented strain expressed Cpa at a higher level than WT BAA-894 (Fig. 3, lanes 1 and 3), which could explain the higher level of serum resistance exhibited by BAA994Δcpa/pMMB66EH::cpa compared to WT BAA-894.
Also, E. coli DH5α transformed with pMMB66EH::cpa expressed Cpa at a higher level than WT BAA-894 (Fig. 3, lanes 1 and 5). This construct was used to determine whether Cpa alone is sufficient to mediate the serum resistance. After 1 h of incubation, 38 and 0.2% of DH5α/pMMB66EH::cpa survived in the presence of 12 and 25% NHS, respectively (Fig. 4A). In contrast, DH5α and DH5α carrying the vector pMMB66EH was rapidly killed by 12 and 25% NHS, but not by HIS (data not shown), indicating that Cpa was able to impart serum resistance to E. coli expressing C. sakazakii Cpa protein. These results support our hypothesis that Cpa plays a role in the serum resistance of C. sakazakii. Furthermore, the higher level of serum resistance of BAA894Δcpa/pMMB66EH::cpa than DH5α/pMMB66EH::cpa suggests that, in addition to Cpa, there are additional Cronobacter factors that may be involved in serum resistance.
Complement pathways involved.
The complement cascade is a complex system of 25 to 30 plasma proteins and plays an innate role in the resistance against microbial infections (31, 45). Three pathways can activate the complement system: the classical, the alternative, and the lectin dependent. To determine which complement activation pathways are involved in Cpa-mediated serum resistance, WT BAA-894 and BAA894Δcpa were incubated with 25% of classical and lectin-dependent complement pathway-inactivated serum (CCPIS) or 25% of alternative complement pathway-inactivated serum (ACPIS). Both the WT and the Cpa mutant strains were resistant to CCPIS. However, BAA894Δcpa was more sensitive than WT BAA-894 to ACPIS (Fig. 4B), suggesting that Cpa is required for resistance to complement and the resistance is activated selectively by the classical and/or lectin pathways.
Proteolytic cleavage of complement components.
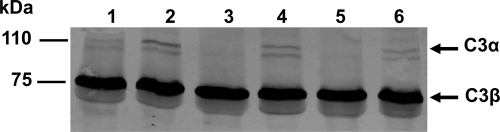
Component C3 plays a central role in the three complement activation pathways (31, 45). In order to investigate whether Cpa is involved in serum resistance by cleaving C3, NHS was incubated with WT BAA-894 and recombinant C. sakazakii and E. coli strains expressing Cpa and then analyzed by immunoblotting with anti-human C3 antibody (Fig. 5). Human C3 is comprised of α and β chains of 110 and 75 kDa, respectively (47). Immunoblot analysis of NHS treated with the WT BAA-894 or one of the Cpa-negative strains, BAA894Δcpa, or DH5α/pMMB66EH, showed intact 110-kDa α- and 75-kDa β-chains of C3 (Fig. 5, lanes 1, 2, and 4), whereas the 110-kDa α-chain band disappeared in NHS incubated with BAA894Δcpa/pMMB66EH::cpa and DH5α/pMMB66EH::cpa (Fig. 5, lanes 3 and 5). These data suggest that overexpression of Cpa resulted in the proteolytic cleavage of the C3 α-chain.
FIG. 5.
Proteolysis of the complement factor C3α chain in NHS by C. sakazakii and recombinant DH5α strains expressing Cpa. 25% NHS was incubated with the following: lane 1, BAA-894; lane 2, BAA894Δcpa; lane 3, BAA894Δcpa/pMMB66EH::cpa; lane 4, DH5α/pMMB66EH; lane 5, DH5α/pMMB66EH::cpa; and lane 6, PBS. C3 proteolytic cleavage was identified by immunoblotting with anti-human C3 antibodies.
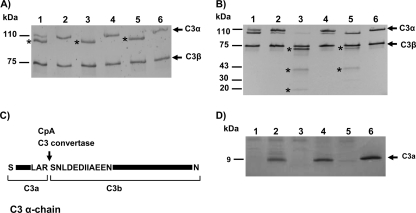
In order to determine whether strains expressing Cpa directly cleave C3, purified C3 was incubated with the bacterial cells followed by SDS-PAGE and immunoblot analysis with anti-human C3 antibodies. After 45 min of incubation at 37°C, the α-chain band was observed at a position (∼100 kDa) lower than that of the intact C3 when the protein was incubated with Cpa-expressing strains BAA994Δcpa/pMMB66EH::cpa and DH5α/pMMB66EH::cpa, suggesting proteolysis of the α-chain band (Fig. 6A, lanes 3 and 5). The 110-kDa α-chain and 100-kDa bands were observed with WT BAA-894, suggesting that these strains caused partial proteolysis of the α-chain band (Fig. 6A, lane 1), compared to the reaction mixture of intact C3 with the cpa mutant where the α-chain band remained unaltered (Fig. 6A, lane 2). After 18 h of incubation at 37°C, further proteolytic cleavage of C3 was observed in the presence of either BAA894Δcpa/pMMB66EH::cpa or DH5α/pMMB66EH::cpa, but not with WT BAA-894 or the cpa mutant strains (Fig. 6B). In addition to the 100-kDa protein, DH5α/pMMB66EH::cpa yielded proteins of approximately 60 and 43 kDa (Fig. 6B, lane 5), whereas the 100-kDa protein was completely degraded to yield proteins of sizes of ca. 60, 43, and 20 kDa when C3 was incubated with BAA994Δcpa/pMMB66EH::cpa (Fig. 6B, lane 3). These results showed that BAA994Δcpa/pMMB66EH::cpa could proteolytically cleave the 110-kDa C3α chain protein better than DH5α/pMMB66EH::cpa.
FIG. 6.
Proteolysis of purified complement factor C3 and C3a by C. sakazakii strains expressing Cpa. (A) Purified C3 was incubated for 45 min with the following: lane 1, BAA-894; lane 2, BAA894Δcpa; lane 3, BAA894Δcpa/pMMB66EH::cpa; lane 4, DH5α/pMMB66EH; lane 5, DH5α/pMMB66EH::cpa; and lane 6, PBS. (B) Purified C3 was incubated for 18 h (lanes are as described for panel A). Proteolytic cleavage of C3 was identified by immunoblotting with anti-human C3 antibodies. Major Cpa-mediated cleavage products of C3 are indicated by asterisks. (C) Schematic view of C3 cleavage site by Cpa after 45 min of incubation. The arrow shows the cleavage site. (D) Purified C3a was incubated with the following: lane 1, BAA-894; lane 2, BAA894Δcpa; lane 3, BAA894Δcpa/pMMB66EH::cpa; lane 4, DH5α/pMMB66EH; lane 5, DH5α/pMMB66EH::cpa; and lane 6, PBS for 18 h. C3a proteolytic cleavage was detected by SDS-PAGE using an 8 to 25% gradient gel. The gel was stained with Coomassie blue.
During activation of the complement, C3 convertase catalyzes the proteolytic cleavage of C3α into C3b and C3a of sizes 101 and 9 kDa, respectively (47). The ∼100-kDa band resulting from the cleavage of C3 by Cpa-expressing strains (Fig. 6A, lanes 1 and 3) appears to be equivalent in size to the C3b α-chain. N-terminal amino acid sequence analysis of this band showed the same C3α cleavage pattern for both CpA and C3 convertase (Fig. 6C). Further, SDS-PAGE analysis showed that WT BAA-894 and Cpa-expressing BAA894Δcpa/pMMB66EH::cpa and DH5α/pMMB66EH::cpa, but not the mutant BAA894Δcpa, degraded C3a (Fig. 6D).
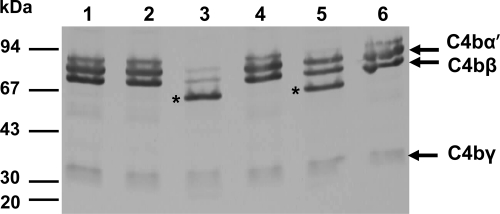
Complement component C4 is the second most abundant component of the complement system and is crucial for activation of both classical and lectin pathways (31, 45). C4b molecule is composed of three disulfide-linked polypeptides chains C4 α′, β, and γ (1). Western blot analysis showed that the Cpa-expressing strain BAA894ΔcpA/pMMB66EH::cpa completely degraded the C4b α′-chain and partially degraded C4b β-chain, resulting in a protein band of ∼60 kDa, whereas the wild-type and cpa mutant strains did not cleave C4b (Fig. 7, lanes 1, 2, and 3). In addition to the C4 α′, β, and γ bands, a 60-kDa protein band was observed when C4b was incubated with DH5α/pMMB66EH::cpa, suggesting a partial proteolysis of the C4 α′ and/or C4 β chains (Fig. 7, lane 5).
FIG. 7.
Proteolysis of purified complement factor C4b by C. sakazakii strains expressing Cpa. Purified C4b was incubated with BAA-894 (lane 1), BAA894Δcpa (lane 2), BAA894Δcpa/pMMB66EH::cpa (lane 3), DH5α/pMMB66EH (lane 4), DH5α/pMMB66EH::cpa (lane 5), and PBS (lane 6) for 18 h. C4b proteolytic cleavage was identified by immunoblotting with anti-human C4 antibodies. Major Cpa-mediated cleavage products of C4b are indicated with asterisks.
Plasminogen activation.
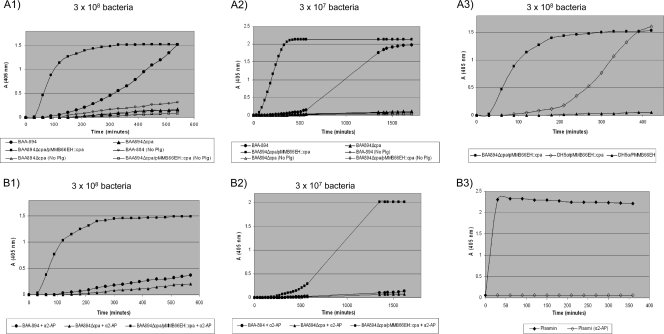
We studied the ability of Cpa to convert plasminogen to plasmin and to inactivate the plasmin inhibitor α2-AP. Our previous results indicated that the proteolytic activity of Cpa on complement components is influenced by Cpa expression levels. To test whether Cpa at different expression levels could activate plasminogen, WT BAA-894, BAA894Δcpa, and BAA894Δcpa/pMMB66EH::cpa were evaluated using different bacterial cell concentrations. We observed that, in the presence of 3 × 108 cells of Cpa-overexpressing strain BAA894Δcpa/pMMB66EH::cpa, there was a rapid activation of plasminogen compared to WT BAA-894, which had a slow plasmin formation, whereas no plasminogen activity was detected for the Cpa mutant strain BAA894Δcpa (Fig. 8A1). No activity was detected when WT BAA-894 or BAA894Δcpa/pMMB66EH::cpa was incubated in the absence of plasminogen (Fig. 8A1), indicating that the plasminogen activity detected in these strains was not due to direct hydrolysis of the chromogenic substrate S-2224 by CpA. Plasminogen activity of BAA-894 and BAA894Δcpa/pMMB66EH::cpa decreased when plasminogen was incubated with 3 × 107 bacteria (Fig. 8A2), showing that plasminogen activity depends on Cpa concentration. In line with the results observed from the serum resistance assay and the proteolysis analysis of complement components C3 and C4b, the pMMB66EH::cpa recombinant plasmid expressed in BAA894Δcpa produced greater plasminogen activity than when it was expressed in E. coli DH5α (Fig. 8A3). This result suggested that Cpa is proteolytically more active when it is expressed in its native C. sakazakii than when it is heterologously expressed in E. coli.
FIG. 8.
Plasminogen activation and α2-AP inactivation by C. sakazakii BAA-894 and recombinant C. sakazakii and E. coli DH5α strains. Bacteria and plasminogen were incubated in PBS without (A) or with (B) α2-AP, and plasmin activity was measured with the chromogenic plasmin substrate S-2251 as described in Materials and Methods. The assays were repeated three times; means from a representative assay with triplicate samples are shown.
We observed that the plasmin inhibitor, α2-AP, did not inhibit the plasmin activity induced by Cpa-overexpressing strain, BAA894Δcpa/pMMB66EH::cpa, in the presence of 3 × 108 bacterial cells and only transiently inhibited the plasmin activity in the presence of 3 × 107 cells (Fig. 8B1 and B2). In contrast, α2-AP permanently inactivated free plasmin and plasmin activity of WT BAA-894 in the presence of both 3 × 108 and 3 × 107 bacteria (Fig. 8B1, B2, and B3). These results indicate that the proteolytic inactivation of α2-AP is also dependent upon the concentration of Cpa.
DISCUSSION
The success of the opportunistic pathogen Cronobacter spp. to cause meningitis is dependent upon its ability to survive in blood and the subsequent invasion of the central nervous system by breaching the blood-brain barrier. In order to achieve these objectives, the bacterial cells must be resistant to complement-dependent killing and must degrade fibrin, collagen, and other structural components of the extracellular matrix (28, 30). Pla and PgtE are able to activate plasminogen to plasmin and inactivate the plasmin inhibitor α2-AP (27, 29, 56). Plasmin is a broad-specificity serine protease that degrades fibrin and collagen, in addition to other structural proteins (41). Plasmin also activates other proteolytic enzymes, such as matrix metalloproteinases (MMPs) that degrade the tight junction components of microvascular endothelial cells (28). This latter function is critical for plasmin-mediated mechanisms of intercellular migration that allow passage of bacterial cells across the vasculature into either peripheral tissues or otherwise privileged compartments such as the central nervous system. Inactivation of α2-AP promotes uncontrolled proteolysis contributing to the invasion of the bacteria (26, 28, 29, 49, 55). Pla and PgtE also mediate serum resistance by proteolytic cleavage of complement components (44, 55).
In the present study, we found that Cpa of C. sakazakii, just like PgtE and Pla, provides resistance against the bactericidal activity of serum by cleaving complement components C3 and C4b, as well as the activation of plasminogen and inactivation of α2-AP. Our results showed that the proteolytic activities of Cpa are influenced by the level of its expression. Overexpression of Cpa in recombinant C. sakazakii or E. coli strains increased the efficiency of plasminogen activation, α2-AP inactivation, and cleavage of complement components compared to Cpa expressed by WT BAA-894. The low proteolytic activity identified in the BAA-894 strain might be due to inhibition by the O antigen. It has been reported that a long O-antigen component of a smooth lipopolysaccharide (LPS) sterically inhibits the proteolytic activity of omptin proteins (25, 29). Similar results were observed for PgtE, where PgtE expressed in a smooth S. enterica strain lacked plasminogen activity, but E. coli harboring pgtE cloned in a multicopy plasmid had plasminogen activity (56). Y. pestis is genetically rough and possesses a short-chain LPS (54); thus, Pla-mediated proteolytic functions are fully active in Y. pestis.
Interestingly, our results showed that even though C. sakazakii cpa mutant and DH5α strains harboring recombinant vector plasmid pMMB66EH::cpa expressed Cpa at the same levels (Fig. 3), BAA894Δcpa/pMMB66EH::cpa was more resistant to NHS and had higher proteolytic activity toward complement components C3 and C4b, plasminogen, and α2-AP than the activity observed with DH5α/pMMB66EH::cpa. It is possible that structural differences in the LPS expressed by C. sakazakii BAA-894 and E. coli allowed Cpa expressed in C. sakazakii to be more proteolytically active than when it is expressed in E. coli. LPS has a dual function on omptins; these proteins need to interact with lipid A to be proteolytically active but also the presence of O-antigen repeats sterically hinders access of exogenous macromolecular substrates to the omptin active site (25, 29). It is known that natural LPS molecules are intrinsically heterogeneous in structure and molecular mass (e.g., differences in oligosaccharide chain lengths and the levels of acylation and substitution) (58). Recent studies have demonstrated that Pla and LPS interact and consequently Pla activity is enhanced by the presence of low amounts of aminoarabinose and low acylation levels of LPS (58).
Our results clearly showed that WT BAA-894 is more resistant to serum killing than the cpa mutant strain. These results suggest that the proteolytic activity of Cpa expressed in WT BAA-894 increases the resistance of the bacteria to serum. Moreover, C. sakazakii is an opportunistic pathogen that causes meningitis, especially in neonates and infants. Neonates do not have a mature immune system and are often unable to mount an effective immune response. The levels of complement components, including C3 and C4, in neonatal plasma are limited compared to those in adults, and generally range from ∼10 to 70% of the adult levels (9). The level of Cpa activity expressed by WT BAA-894 would be adequate to afford resistance to complement mediated-killing given that neonatal levels of serum complement components are significantly less than that of an adult.
It is also possible that during infection with C. sakazakii, the LPS structure and CpA expression are modified in such way that the activity of Cpa is maximized. In fact, during growth of S. enterica within mouse macrophages, the LPS structure is altered and the length and the amount of the O antigen are strongly reduced (6, 29). The same studies also showed that the expression levels of PgtE are increased during growth of the bacteria inside of the macrophage. Recently, Suomalainen et al. (58) showed that temperature induces changes in Y. pestis LPS, which in turn would lead to an increase in Pla-mediated proteolysis. Studies using in vivo models are necessary to further determine the role of Cpa in the virulence of C. sakazakii.
On the other hand, we showed that even though E. coli recombinant strain DH5α/pMMB66EH::cpa cleaves complement components C3 and C4b more efficiently than WT BAA894, this strain is more resistant to NHS. These results suggest that in addition to Cpa there are other Cronobacter factors involved in serum resistance. This is further supported by our results showing that BAA894Δcpa is more resistant to NHS than DH5α. Mittal et al. (36), using neonatal rat serum, found that the outer membrane protein OmpA is necessary for the survival of Cronobacter spp. in serum.
Complement component C3 plays a central role in the activation of the complement system (47). Its activation is required for classical, lectin, and alternative pathways. In all three pathways, a C3-convertase cleaves C3 into C3a and C3b. C3b binds to the surface of the pathogen, causing a cascade of further complement protein cleavage and activation events that culminate in the formation of the membrane attack complex. C3b binding to the surface of pathogens also leads to greater internalization of a pathogen by phagocytic cells via opsonization. Our data demonstrate that Cpa proteolytically cleaves C3 α-chain when it is incubated with NHS or purified C3 for 18 h. Similar activity has been reported for the elastase of Pseudomonas aeruginosa (17, 59). This protease was shown to degrade the C3 α-chain without affecting the C3 β-chain.
Interestingly, incubation of Cpa-expressing C. sakazakii and recombinant DH5α strains with purified C3 for only 45 min cleaved C3α-chain protein at the same site where C3 convertase cleaves C3 α protein to produce C3a and C3b. This activity of Cpa against C3 is reminiscent of that of gelatinase, GelE, produced by Enterococcus faecalis (40). Purified GelE cleaved C3 into a C3b-like molecule, which was inactivated rapidly via reaction with water. This C3 convertase-like activity of GelE was shown to result in the depletion of C3, thus inhibiting the activation of the complement system (40). Alternatively, it is possible that the presence of Cpa in BAA-894 and recombinant C. sakazakii and E. coli strains induces a spontaneous cleavage of C3 in a short period of time to produce C3b and C3a. To determine whether Cpa induces this spontaneous or the C3b 100-kb band is produced by Cpa proteolytic activity, it would be necessary to incubate purified C3 with a C. sakazakii strain expressing a proteolytic inactivated Cpa.
Our results also indicate that Cpa expressed by WT BAA-894 or recombinant C. sakazakii and E. coli DH5α completely degrades the small-molecular-weight C3a protein. C3a is an anaphylatoxin that serves as a mediator of inflammation by inducing mast cell degranulation, histamine release, and increased vascular permeability (31, 45). Degradation of C3a by Cpa may cause paralysis of a variety of C3a-mediated immune reactions occurring in the human serum.
Our finding also showed that inactivation of the classic and lectin pathway of the complement restored viability of cpa mutant at the same level as the WT strain. Furthermore, inactivation of the alternative pathway did not restore the viability of the cpa mutant, indicating that the cpa mutant is killed by a classical/lectin pathway only and thus demonstrating that Cpa is required for exerting resistance to complement activation by the classical and/or lectin pathways. Activation of the classical pathway is initiated by the binding of C1 complex to antibodies bound to an antigen on the bacterial surface. C1s, in the complex, activates C4 by cleaving C4 to C4a and C4b, which bind to the microbial surface (3, 8, 19). Our results indicate that Cpa provides resistance to the classical complement pathway by proteolytically cleaving C4b α and C4 β-chains (Fig. 7). We do not rule out the possibility that in addition to cleaving C3, C3a, and C4b, Cpa may proteolytically degrade other components of the complement cascade, such as C5.
In conclusion, we have shown that Cpa expressed by WT C. sakazakii BAA-894 slowly cleaves plasminogen and increases survival in serum in comparison to a Cpa deletion mutant. The low proteolytic activity of BAA-894 may be due to Cpa inhibition by BAA-894 smooth LPS. Overexpression of Cpa in recombinant C. sakazakii produces a rapid activation of plasminogen and inactivation of α2-AP, as well as enhances proteolytic cleavage of complement components that in turn increase resistance to serum bactericidal activity. In vivo studies are necessary to further determine the role of Cpa in the virulence of C. sakazakii.
Acknowledgments
We thank Carmen Fernandez-Prada, Cynthia L. Sears, and Prasad Rallabhandi for reviewing the manuscript.
L.H. is an FDA Commissioner's Fellow. K.G.J. and C.J.G. are Oak Ridge Institute for Science and Education fellows, and we thank the Department of Energy for their support.
Editor: B. A. McCormick
Footnotes
Published ahead of print on 18 January 2011.
REFERENCES
- 1.Chan, A. C., and J. P. Atkinson. 1985. Oligosaccharide structure of human C4. Immunol. 134:1790-1798. [PubMed] [Google Scholar]
- 2.Clark, N. C., B. C. Hill, C. M. O'Hara, O. Steingrimsson, and R. C. Cooksey. 1990. Epidemiologic typing of Enterobacter sakazakii in two neonatal nosocomial outbreaks. Diagn. Microbiol. Infect. Dis. 13:467-472. [DOI] [PubMed] [Google Scholar]
- 3.Clay, C. D., S. Soni, J. S. Gunn, and L. S. Schlesinger. 2008. Evasion of complement-mediated lysis and complement C3 deposition are regulated by Francisella tularensis lipopolysaccharide O antigen. J. Immunol. 181:5568-5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Donnenberg, M. S., and J. B. Kaper. 1991. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect. Immun. 59:4310-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Egile, C., H. d'Hauteville, C. Parsot, and P. J. Sansonetti. 1997. SopA, the outer membrane protease responsible for polar localization of IcsA in Shigella flexneri. Mol. Microbiol. 23:1063-1073. [DOI] [PubMed] [Google Scholar]
- 6.Eriksson, S., S. Lucchini, A. Thompson, M. Rhen, and J. C. Hinton. 2003. Unraveling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol. Microbiol. 47:103-118. [DOI] [PubMed] [Google Scholar]
- 7.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 8.Fine, D. P., S. R. Marney, Jr, D. G. Colley, J. S. Sergent, and R. M. DesPrez. 1972. C3 shunt activation in human serum chelated with EGTA. J. Immunol. 109:807-809. [PubMed] [Google Scholar]
- 9.Firth, M. A., P. E. Shewen, and D. C. Hodgins. 2005. Passive and active components of neonatal innate immune defenses. Anim. Health Res. Rev. 6:143-158. [DOI] [PubMed] [Google Scholar]
- 10.Fürste, J. P., et al. 1986. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene 48:119-131. [DOI] [PubMed] [Google Scholar]
- 11.Guina, T., E. C. Yi, H. Wang, M. Hackett, and S. I. Miller. 2000. A PhoP-regulated outer membrane protease of Salmonella enterica serovar Typhimurium promotes resistance to alpha-helical antimicrobial peptides. J. Bacteriol. 182:4077-4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gurtler, B., J. L. Kornacki, and L. R. Beuchat. 2005. Enterobacter sakazakii: a coliform of increased concern to infant health. Int. J. Food Microbiol. 104:1-34. [DOI] [PubMed] [Google Scholar]
- 13.Haiko, J., L. Laakkonen, K. Juuti, N. Kalkkinen, and T. K. Korhonen. 2010. The omptins of Yersinia pestis and Salmonella enterica cleave the reactive center loop of plasminogen activator inhibitor 1. J. Bacteriol. 192:4553-4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haiko, J., M. Suomalainen, T. Ojala, K. Lähteenmäki, and T. K. Korhonen. 2009. Breaking barriers: attack on innate immune defences by omptin surface proteases of enterobacterial pathogens. Innate Immun. 15:67-80. [DOI] [PubMed] [Google Scholar]
- 15.Haiko, J., M. Kukkonen, J. J. Ravantti, B. Westerlund-Wikstrom, and T. K. Korhonen. 2009. The single substitution I259T, conserved in the plasminogen activator Pla of pandemic Yersinia pestis branches, enhances fibrinolytic activity. J. Bacteriol. 191:4758-4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Himelright, I., E. Harris, V. Lorch, and M. Anderson. 2002. Enterobacter sakazakii infections associated with the use of powdered infant formula—Tennessee, 2001. JAMA 287:2204-2205. [PubMed] [Google Scholar]
- 17.Hong, Y. Q., and B. Ghebrehiwet. 1992. Effect of Pseudomonas aeruginosa elastase and alkaline protease on serum complement and isolated components C1q and C3. Clin. Immunol. Immunopathol. 62:133-138. [DOI] [PubMed] [Google Scholar]
- 18.Hritonenko, V., and C. Stathopoulos. 2007. Omptin proteins: an expanding family of outer membrane proteases in Gram-negative Enterobacteriaceae. Mol. Membr. Biol. 24:395-406. [DOI] [PubMed] [Google Scholar]
- 19.James, K. 1982. Complement: activation, consequences, and control. Am. J. Med. Technol. 48:735-742. [PubMed] [Google Scholar]
- 20.Johnson, J. R., B. Johnston, M. A. Kuskowski, R. Colodner, and R. Raz. 2005. Spontaneous conversion to quinolone and fluoroquinolone resistance among wild-type Escherichia coli isolates in relation to phylogenetic background and virulence genotype. Antimicrob. Agents Chemother. 49:4739-4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim, K., et al. 2010. Outer membrane proteins A (OmpA) and X (OmpX) are essential for basolateral invasion of Cronobacter sakazakii. Appl. Environ. Microbiol. 76:5188-5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kleiman, M. B., S. D. Allen, P. Neal, and J. Reynolds. 1981. Meningoencephalitis and compartmentalization of the cerebral ventricles caused by Enterobacter sakazakii. J. Clin. Microbiol. 14:352-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kothary, M. H., B. A. McCardell, C. D. Frazar, D. Deer, and B. D. Tall. 2007. Characterization of the zinc-containing metalloprotease encoded by zpx and development of a species-specific detection method for Enterobacter sakazakii. Appl. Environ. Microbiol. 73:4142-4151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kucerova, E., et al. 2010. Genome sequence of Cronobacter sakazakii BAA-894 and comparative genomic hybridization analysis with other Cronobacter species. PLoS One 5:e9556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kukkonen, M., et al. 2004. Lack of O-antigen is essential for plasminogen activation by Yersinia pestis and Salmonella enterica, Mol. Microbiol. 51:215-225. [DOI] [PubMed] [Google Scholar]
- 26.Kukkonen, M., and T. K. Korhonen. 2004. The omptin family of enterobacterial surface proteases/adhesins: from housekeeping in Escherichia coli to systemic spread of Yersinia pestis. Int. J. Med. Microbiol. 294:7-14. [DOI] [PubMed] [Google Scholar]
- 27.Kukkonen, M., et al. 2001. Protein regions important for plasminogen activation and inactivation of α2-antiplasmin in the surface protease Pla of Yersinia pestis. Mol. Microbiol. 40:1097-1111. [DOI] [PubMed] [Google Scholar]
- 28.Lahteenmaki, K., S. Edelman, and T. K. Korhonen. 2005. Bacterial metastasis: the host plasminogen system in bacterial invasion. Trends Microbiol. 13:79-85. [DOI] [PubMed] [Google Scholar]
- 29.Lähteenmäki, K., P. Kyllönen, L. Partanen, and T. K. Korhonen. 2005. Anti-protease inactivation by Salmonella enterica released from infected macrophages. Cell Microbiol. 7:529-538. [DOI] [PubMed] [Google Scholar]
- 30.Lahteenmaki, K., M. Kukkonen, and T. K. Korhonen. 2001. The Pla surface protease/adhesin of Yersinia pestis mediates bacterial invasion into human endothelial cells. FEBS Lett. 504:69-72. [DOI] [PubMed] [Google Scholar]
- 31.Law, S. K. A., and K. B. M. Reid. 1995. Complement, 2nd ed. IRL Press, Oxford, United Kingdom.
- 32.Lehner, A., and R. Stephan. 2004. Microbiological, epidemiological, and food safety aspects of Enterobacter sakazakii, J. Food Prot. 67:2850-2857. [DOI] [PubMed] [Google Scholar]
- 33.Mangel, W. F., et al. 1994. Omptin: an Escherichia coli outer membrane proteinase that activates plasminogen. Methods Enzymol. 244:384-399. [DOI] [PubMed] [Google Scholar]
- 34.McCarter, J. D., et al. 2004. Substrate specificity of the Escherichia coli outer membrane protease OmpT. J. Bacteriol. 186:5919-5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Merino, S., S. Albertí, and J. M. Tomás. 1994. Aeromonas salmonicida resistance to complement-mediated killing. Infect. Immun. 62:5483-5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mittal, R., Y. Wang, C. J. Hunter, I. Gonzalez-Gomez, and N. V. Prasadarao. 2009. Brain damage in newborn rat model of meningitis by Enterobacter sakazakii: a role for outer membrane protein A. Lab. Invest. 89:263-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mohan Nair, M. K., and K. Venkitanarayanan. 2007. Role of bacterial OmpA and host cytoskeleton in the invasion of human intestinal epithelial cells by Enterobacter sakazakii. Pediatr. Res. 62:664-669. [DOI] [PubMed] [Google Scholar]
- 38.Nazarowec-White, M., and J. M. Farber. 1997. Enterobacter sakazakii: a review. Int. J. Food Microbiol. 34:103-113. [DOI] [PubMed] [Google Scholar]
- 39.Pagotto, F. J., M. Nazarowec-White, S. Bidawid, and J. M. Farber. 2003. Enterobacter sakazakii: infectivity and enterotoxin production in vitro and in vivo. J. Food Prot. 66:370-375. [DOI] [PubMed] [Google Scholar]
- 40.Park, S. Y., et al. 2008. Immune evasion of Enterococcus faecalis by an extracellular gelatinase that cleaves C3 and iC3b. J. Immunol. 181:6328-6336. [DOI] [PubMed] [Google Scholar]
- 41.Pointing, C. P., J. M. Marshall, and S. A. Cederholm-Williams. 1992. Plasminogen: a structural review. Blood Coagul. Fibrinolysis 3:605-614. [PubMed] [Google Scholar]
- 42.Raghav, M., and P. K. Aggarwal. 2007. Purification and characterization of Enterobacter sakazakii enterotoxin. Can. J. Microbiol. 53:750-755. [DOI] [PubMed] [Google Scholar]
- 43.Ramu, P., et al. 2008. Activation of pro-matrix metalloproteinase-9 and degradation of gelatin by the surface protease PgtE of Salmonella enterica serovar Typhimurium. Int. J. Med. Microbiol. 298:263-278. [DOI] [PubMed] [Google Scholar]
- 44.Ramu, P., et al. 2007. The surface protease PgtE of Salmonella enterica affects complement activity by proteolytically cleaving C3b, C4b, and C5. FEBS Lett. 581:1716-1720. [DOI] [PubMed] [Google Scholar]
- 45.Rautemaa, R., and S. Meri. 1999. Complement-resistance mechanisms of bacteria. Microbes Infect. 1:785-794. [DOI] [PubMed] [Google Scholar]
- 46.Rhee, K. J., et al. 2009. Induction of persistent colitis by a human commensal, enterotoxigenic Bacteroides fragilis, in wild-type C57BL/6 mice. Infect. Immun. 77:1708-1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sahu, A., and J. D. Lambris. 2001. Structure and biology of complement protein C3, a connecting link between innate and acquired immunity. Immunol. Rev. 180:35-48. [DOI] [PubMed] [Google Scholar]
- 48.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 49.Sebbane, F., C. O. Jarrett, D. Garner, D. Long, and B. J. Hinnebush. 2006. Role of the Yersinia pestis plasminogen activator in the incidence of distinct septicemic and bubonic forms of flea-borne plague. Proc. Natl. Acad. Sci. U. S. A. 103:5526-5530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shere, K. D., S. Sallustio, A. Manessis, T. G. D'Aversa, and M. B. Goldberg. 1997. Disruption of IcsP, the major Shigella protease that cleaves IcsA, accelerates actin-based motility. Mol. Microbiol. 25:451-462. [DOI] [PubMed] [Google Scholar]
- 51.Simmons, B. P., M. S. Gelfans, M. Haas, L. Metts, and J. Ferguson. 1989. Enterobacter sakazakii infections in neonates associated with intrinsic contamination of a powdered infant formula. Infect. Control Hosp. Epidemiol. 10:398-401. [DOI] [PubMed] [Google Scholar]
- 52.Simon, R., U. Priefer, and A. Pülher. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Biotechnology (NY) 1:784-791. [Google Scholar]
- 53.Singamsetty, V. K., Y. Wang, H. Shimada, and N. V. Prasadarao. 2008. Outer membrane protein A expression in Enterobacter sakazakii is required to induce microtubule condensation in human brain microvascular endothelial cells for invasion. Microb. Pathog. 45:181-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Skurnik, M., A. Peippo, and E. Ervela. 2000. Characterization of the O-antigen gene cluster of Yersinia pseudotuberculosis and the cryptic O-antigen gene cluster of Yersinia pestis shows that the plague bacillus is most closely related to and has evolved from Y. pseudotuberculosis serotype O:1b. Mol. Microbiol. 37:316-330. [DOI] [PubMed] [Google Scholar]
- 55.Sodeinde, O. A., et al. 1992. A surface protease and the invasive character of plague. Science 258:1004-1007. [DOI] [PubMed] [Google Scholar]
- 56.Sodeinde, O. A., and J. D. Goguen. 1989. Nucleotide sequence of the plasminogen activator gene of Yersinia pestis: relationship to ompT of Escherichia coli and gene E of Salmonella typhimurium, Infect. Immun. 57:1517-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stumpe, S., R. Schmid, D. L. Stephens, G. Georgiou, and E. P. Bakker. 1998. Identification of OmpT as the protease that hydrolyzes the antimicrobial peptide protamine before it enters growing cells of Escherichia coli. J. Bacteriol. 180:4002-4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Suomalainen, M., et al. 2010. Temperature-induced changes in the lipopolysaccharide of Yersinia pestis affect plasminogen activation by the pla surface protease. Infect. Immun. 78:2644-2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Suter, S., U. B. Schaad, L. Roux, U. E. Nydegger, and F. A. Waldvogel. 1984. Granulocyte neutral proteases and Pseudomonas elastase as possible causes of airway damage in patients with cystic fibrosis. J. Infect. Dis. 149:523-531. [DOI] [PubMed] [Google Scholar]
- 60.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 61.van Acker, J., et al. 2001. Outbreak of necrotizing enterocolitis associated with Enterobacter sakazakii in powdered milk formula. J. Clin. Microb. 39:293-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vandeputte-Rutten, L., et al. 2001. Crystal structure of the outer membrane protease OmpT from Escherichia coli suggests a novel catalytic site. EMBO J. 20:5033-5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zuckerkandl, E., and L. Pauling. 1965. Evolutionary divergence and convergence in proteins, p. 97-166. In V. Bryson and H. J. Vogel (ed.), Evolving genes and proteins. Academic Press, Inc., New York, NY.