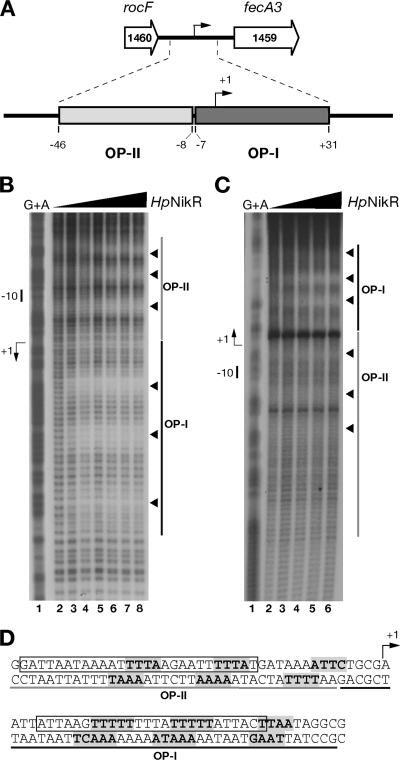
FIG. 1.
Hydroxyl radical footprinting of Ni-HpNikR on the PfecA3 region. (A) The fecA3 gene of H. pylori G27 (HPG27_1459, nucleotides 1586410 to 1583885, minus strand) is separated by 352 nucleotides from the upstream gene rocF (HPG27_1460, nucleotides 1587730 to 1586762, minus strand). Two operator sites for HpNikR were mapped by DNase I footprinting within this promoter: operator I (OP-I, dark gray), spanning nucleotides −7 to +31, and operator II (OP-II, light gray), spanning nucleotides −46 to −8. (B and C) DNA probes comprising the fecA3 promoter were challenged with increasing concentrations of HpNikR in a hydroxyl radical footprinting assay and separated on a denaturing sequencing gel. Approximately 20 fmol of 5′-end-labeled probes was incubated with increasing concentrations of protein (HpNikR tetramer): 0 nM (lanes 1), 24.7 nM (lanes 2), 49 nM (lanes 3), 112.7 nM (lanes 4), 245 nM (lanes 5), 490 nM (lanes 6), 980 nM (lane 7), and 1,960 nM (lane 8). A G+A sequence reaction ladder of the each probe was run in parallel to map the bound bases with respect to the transcriptional start (+1) indicated by a bent arrow on the left side of each panel. The relative positions of OP-I and -II are represented by dark and light gray bars, respectively, and the short nucleotide segments protected by HpNikR from hydroxyl radical cuts are shown by arrowheads on the right side. The separation gels were run at 54 W for a variable time (75 min to 3 h 15 min) to resolve the protected bases on each strand, and representative gels are shown for the noncoding (B) and coding (C) strands. Since the original cloning strategy placed the HpNikR operators at about 50 bp in one case and at over 150 bp in the other with respect to the 5′-labeled ends, the base separation is better resolved along the noncoding strand than on the coding one. (D) Nucleotide sequences of the fecA3 operators. The bases directly contacted by HpNikR are shown in bold and shaded in gray. Three short stretches are protected along both OP-I and OP-II; the bent arrow shows the transcriptional start site. The sequences comprising OP-II and OP-I are underlined in gray and black, respectively. The sequence containing the consensus motif originally proposed by Delany et al. and the revised consensus proposed by Stoof et al. (15, 37) are boxed.