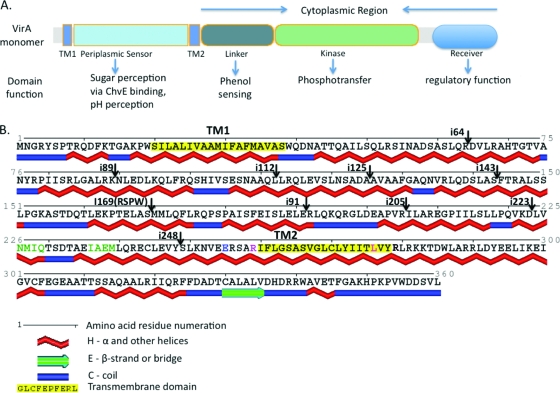
FIG. 1.
(A) Schematic depiction of VirA domain organization in a VirA monomer and known functions of each domain. Two transmembrane domains, TM1 and TM2 (amino acids highlighted in yellow in panel B), delineate the periplasmic domain. The cytoplasmic domain contains the sites of phenol perception (linker) and the kinase domain that has a histidine residue essential for phosphotransfer at position 474. Note that the receiver domain, though assigned an inhibitory role by Chang and Winans (15), has recently been found to have a positive regulatory effect on VirA kinase activity (72). (B) Predicted secondary structure of the N-terminal region of wild-type VirA. Secondary structure predictions were made from the amino acid sequence by using SABLE (1, 2, 68) and rendered using Polyview (60, 61). Note the predominantly α-helical nature of this region and the sites of different four-amino-acid insertions used in this study. Residues in green signify a small motif identical to a periplasmic binding protein interaction motif found in the E. coli Trg chemoreceptor. Amino acids N226, Q229, and I236 were the subjects of a site-directed mutagenesis study reported here to evaluate the importance of this motif in signal integration by VirA. A previously reported E255-to-L amino acid mutation that causes a loss of AS sensitivity (5) and was used in this study is also depicted (residue in blue). Also shown here are the positions where four helix-breaking amino acids were inserted into various predicted α-helices (shown by arrows pointing down), and the two amino acids, R259 and L276 (depicted in red), that were replaced by a serine and arginine, respectively, in this study. The R259S/L276R double mutation results in a TM2 domain that ends at position 276 instead of 278, according to prediction programs.