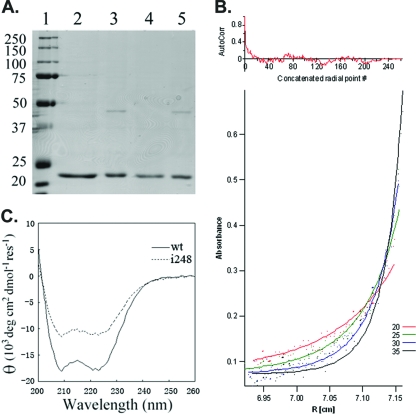
FIG. 3.
VirAperi isolation, dimerization, and helical abundance. (A) SDS-PAGE (10%) analysis of the wild-type VirAperi and the insertion mutant (i248). Lane 1, protein standards with molecular sizes labeled in kDa; lanes 2 and 3, wild-type VirAperi before and after dialysis against buffer A (20 mM HEPES, pH 8.5, 50 mM NaCl), respectively; lanes 4 and 5, VirAperi with the i248 mutation before and after dialysis against buffer A, respectively. The elution buffer for purification is 20 mM HEPES, pH 8.5, 500 mM NaCl, 40 mM DTT, and 1 mM EDTA. (B) Analytical ultracentrifugation analysis of VirAperi. The dots in the lower panel represent measured absorbances at 280 nm taken at 20,000, 25,000, 30,000, and 35,000 rpm. The best-fit curves (continuous lines) for the equilibrium model in a monomer-dimer equilibrium with a Kd of 39.8 μM are superimposed. AutoCorr, autocorrelation; R, radius. (C) Far-UV CD spectroscopy of wild-type (wt) VirAperi and VirAperi with i248. The spectra of both wt VirAperi and VirAperi with i248 were obtained at a concentration of 7.5 μM in 50 mM NaHPO4 buffer (pH 7.6) and 50 mM NaCl. Measurements were taken in 1-nm increments from 260 to 200 nm in a 0.1-cm-path-length cuvette with a bandwidth of 1 nm on a Jasco J810 spectropolarimeter. The spectrum was the average of three scans at 20°C. res, residue.