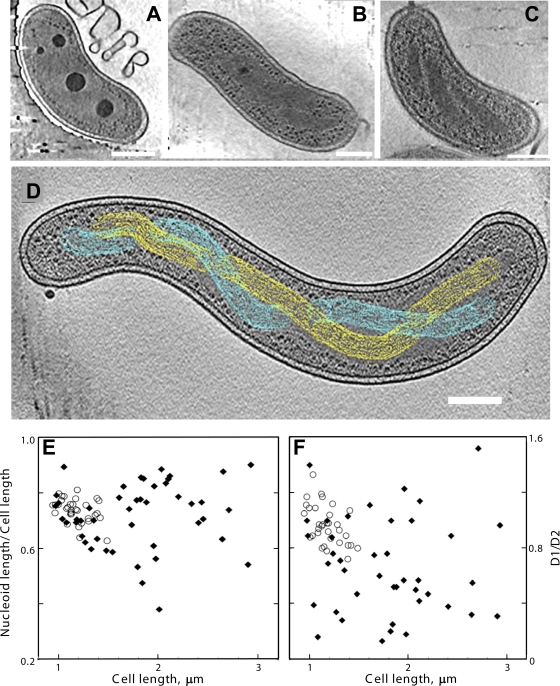
FIG. 1.
Cryo-electron tomography of HD100 wild-type and MreB2-mTFP B. bacteriovorus cells. (A) Tomographic slice (12.5 nm thick) of a wild-type cell that has a compact nucleoid surrounded by dark granular structures, presumably bacterial ribosomes. (B) Tomographic slice (12.5 nm thick) of a wild-type cell that has its nucleoid organized in two twisted strands. The strand thickness is ∼75 nm, with a helical repeat that varies from 320 nm to 334 nm along the cell length. (C) Tomographic slice (21 nm thick) of an MreB2-mTFP cell that has distinct nucleoid helical banding. (D) Depiction of the path of separated strands (in yellow and cyan) of the spiral nucleoid in an MreB2-mTFP cell where the strands could be resolved along the entire length of the cell. The variation in width of the strand along the spiral can be seen clearly in the cellular tomogram (see Video S3 in the supplemental material); for simplicity, this variation is not highlighted in the colored rendering of the spiral done using the object editor in the program IMOD (28). The volume occupied by one nucleoid strand is indicated using yellow mesh, and blue mesh is used for the volume of the other. (E and F) Nucleoid size plotted as a function of cell length (E) and positioning of the ends of the nucleoids relative to the anterior (distance D1) and posterior (distance D2) cellular poles (F). The ratio of D1/D2 is plotted as a function of cell length. In both plots, distances in wild-type cells are indicated by open circles, and distances in mutant cells are indicated by closed diamonds. Bar, 200 nm.