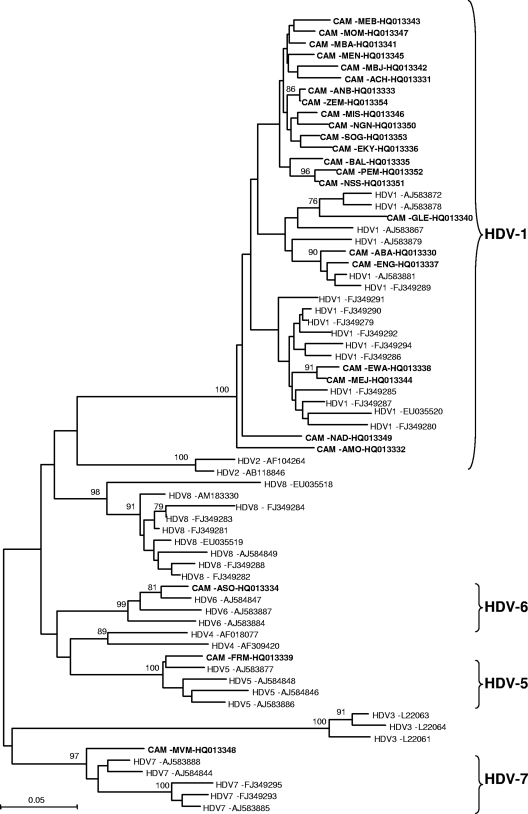
Fig. 1.
Phylogenetic analysis of a 360-bp fragment of the HDV-sHD gene from different HDV strains using the neighbor-joining (NJ) method. Twenty-five HDV sequences from Cameroonian patients (highlighted in boldface; name of the isolate and GenBank accession number are indicated) were aligned with 41 previously published HDV sequences of clades 1 to 8 (HDV-1 to HDV-8) available from GenBank as reference genes (Genotype and GenBank accession number are indicated) by using the CLUSTAL X v1.81 software program and imported into MEGA v.4.0 software for phylogenetic analysis. Numbers next to the nodes of the tree represent bootstrap values (1,000 replicates). Only bootstrap values above 70% are presented. Genetic distances were calculated using the Kimura two-parameter method.