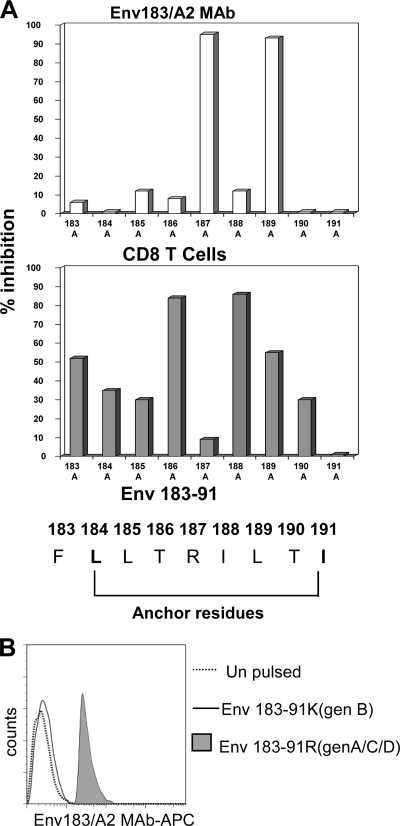
FIG. 4.
Fine-mapping of Env183/A2 MAb epitope recognition. (A) Influence of amino acid mutations within the Env183-91 epitope on Env183/A2 MAb and CD8 T-cell recognition. T2 cells were pulsed with 1 μM Env183-91 peptide with or without (wild type [wt]) alanine substitutions at the indicated positions. The inhibition of TCR-like MAb binding (top) and CD8 T-cell activation (bottom) elicited by alanine substitutions on the wild-type Env183-91 peptide are indicated by bars. Percent inhibition is calculated with the following formula: MFI (or CD107 expression) values obtained with alanine-substituted peptides divided by values obtained with wild-type peptides × 100. (B) Env183/A2 MAb is exclusively specific for the Env183-91 peptide of HBV genotypes (gen) A, C, and D. T2 cells were pulsed with 1 μM Env183-91 peptides with the indicated substitutions at position 187. 187R is characteristic of HBV genotype A, C, and D isolates, and K187 is characteristic of HBV genotype B, E, and F isolates. These cells were stained with Env183/A2 MAb for 1 h and analyzed by flow cytometry. Histograms were overlaid. Results are representative of three independent experiments.