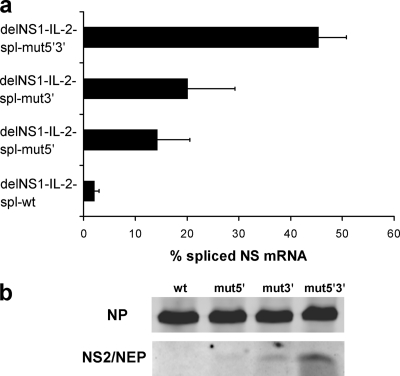
FIG. 3.
(a) Real-time PCR analysis indicating the percentage of spliced NS mRNA in an in vitro replication/transcription system. Vero cells were transfected with bidirectional plasmids (9) coding for the viral RNP proteins and a plasmid coding for the indicated chimeric delNS1-IL-2 virus segment containing either wild-type or mutated splice sequences. After 24 h, RNA was extracted from transfected cells, reverse transcribed, and analyzed for spliced and unspliced NS mRNA steady-state levels by real-time PCR using a SYBR green detection system (Applied Biosystems). For absolute quantitation, suitable serial dilutions of pcDNA-NEP (for spliced mRNA) and pHW-PR8-NS (for unspliced mRNA) were used as standards. All PCRs were run in triplicate. The amplification of single bands was verified by agarose gel electrophoresis and melting curve analysis. The values were calculated as the percentage of spliced NS mRNA and are means ± standard deviations from three independent experiments. (b) NS2/NEP protein expression parallels NS mRNA splicing efficacy. Vero cells were transfected as described for panel a and harvested 24 h posttransfection. Whole-cell extracts were separated by sodium dodecyl sulfate-15% polyacrylamide gel electrophoresis and analyzed by immunoblotting using a polyclonal NS2/NEP-specific antiserum (unpublished data) and, as a control, a blend of two monoclonal NP-specific antibodies (Millipore). Splice site mutations are indicated on the top. wt, wild type.