Abstract
Viremia is significantly lower in HIV-2 than in HIV-1 infection, irrespective of disease stage. Nevertheless, the comparable proviral DNA burdens observed for these two infections indicate similar numbers of infected cells. Here we investigated this apparent paradox by assessing cell-associated viral replication. We found that untreated HIV-1-positive (HIV-1+) and HIV-2+ individuals, matched for CD4 T cell depletion, exhibited similar gag mRNA levels, indicating that significant viral transcription is occurring in untreated HIV-2+ patients, despite the reduced viremia (undetectable to 2.6 × 104 RNA copies/ml). However, tat mRNA transcripts were observed at significantly lower levels in HIV-2+ patients, suggesting that the rate of de novo infection is decreased in these patients. Our data also reveal a direct relationship of gag and tat transcripts with CD4 and CD8 T cell activation, respectively. Antiretroviral therapy (ART)-treated HIV-2+ patients showed persistent viral replication, irrespective of plasma viremia, possibly contributing to the emergence of drug resistance mutations, persistent hyperimmune activation, and poor CD4 T cell recovery that we observed with these individuals. In conclusion, we provide here evidence of significant ongoing viral replication in HIV-2+ patients, further emphasizing the dichotomy between amount of plasma virus and cell-associated viral burden and stressing the need for antiretroviral trials and the definition of therapeutic guidelines for HIV-2 infection.
HIV-2 infection is characterized by a low or undetectable plasma viral load (3, 5, 46, 50, 54), in agreement with its reduced transmission rate (2, 26, 30), providing a natural model to investigate the relative contribution of HIV replication to AIDS progression.
Notwithstanding the two-log difference in viremia levels that characterize HIV-2 and HIV-1 infection, similar levels of cell-associated viral DNA have been reported, suggesting a comparable number of infected cells (6, 24, 45, 52). This apparent paradox highlights the potential contribution of quiescent latent virus to proviral load and the relevance of quantifying the ongoing viral replication. The only study, to our knowledge, addressing transcriptional activity in HIV-2-positive (HIV-2+) patients demonstrated lower gag mRNA expression levels in HIV-2+ than in HIV-1+ individuals (34). However, the fact that these cohorts were not paired for disease stage is a possible confounding factor, as it is known that gag mRNA levels are increased in HIV-1+ individuals with low CD4 T cell counts (22, 39).
In spite of the much lower rate of CD4 T cell decline in HIV-2 than in HIV-1 infection (19, 35), a progressive CD4 T cell depletion, ultimately leading to AIDS, is observed with HIV-2+ individuals (12, 36). Importantly, despite the lack of clinical trials of antiretroviral therapy (ART) in HIV-2 infection, the majority of reports showed poor immunological recovery in ART-treated HIV-2+ patients, even in the context of suppression of viremia (1, 19, 29, 40, 51, 56). A rapid emergence of drug-associated mutations in HIV-2+ patients under ART has also been reported (7, 13-15, 25, 29, 41, 47, 48), suggesting that there is some ongoing viral replication. There are no data regarding the impact of ART on HIV-2 proviral DNA levels and/or HIV-2 transcriptional activity.
We have previously shown that CD4 T cell depletion is directly related to immune activation but only indirectly to plasma viral load in both HIV-2 and HIV-1 infections (28, 55). Here we investigate the relationship of CD4 T cell levels and T cell activation with cell-associated viral burden, measured in terms of gag and tat mRNA and proviral DNA levels, in parallel with plasma viremia in cohorts of untreated HIV-2+ and HIV-1+ individuals together with a cohort of ART-treated HIV-2+ individuals.
MATERIALS AND METHODS
Studied cohorts.
We assessed 45 HIV-2+ (16 of which were receiving ART) and 27 untreated HIV-1+ patients followed at Hospital de Santa Maria in Lisbon, Portugal, as well as 16 HIV-seronegative age-matched controls. None of the patients had ongoing opportunistic infections or tumors. Tables 1 and 2 describe untreated and treated cohorts, respectively. Although HIV-2-infected cohorts included an increased number of women, non-Caucasian, and elderly individuals, in the current study these factors were not found to significantly impact the parameters under analysis (data not shown). All subjects gave informed consent for blood sampling and processing. The study was approved by the Ethical Board of the Faculty of Medicine, University of Lisbon.
TABLE 1.
Characterization of untreated HIV-2+, untreated HIV-1+, and seronegative cohortsc
| Parameter | Seronegative | Untreated HIV-2 | Untreated HIV-1 |
|---|---|---|---|
| No. (male/female) | 16 (6/10) | 29 (9/20) | 27 (18/9) |
| Median age (range) (yr) | 45 (27-57) | 52 (19-78)# | 39 (23-61) |
| Ethnicity (Caucasian/other) (no.) | 15/1 | 15/14 | 20/7 |
| CD4 T cells/μl (range) | 818 (518-1,312) | 568 (52-1,586)* | 372 (18-1,848)** |
| % CD4 T cells (range) | 43 (34-61) | 28 (7-54)*** | 20 (1-47)*** |
| CD8 T cells/μl (range) | 494 (213-1,109) | 788 (271-1,701)** | 1065 (177-2,996)*** |
| % CD8 T cells (range) | 27 (12-42) | 37 (23-74)*** | 50 (16-69)*** |
| Viremia (range) (RNA copies/ml)a | NA | 200 (200-2.6 × 104)### | 1.4 × 104 (40-4.5 × 106) |
| Proviral DNA (range) (copies/106 PBMC)b | NA | 5 (5-1033) | 54 (5-975) |
HIV-2 viremia was below 200 RNA copies/ml (cutoff) in 22 out of 29 patients studied; HIV-1 viremia was below 40 RNA copies/ml (cutoff) in 3 out of 24 patients.
Proviral DNA was below 5 copies/106 PBMC (cutoff) in 15 out of the 29 HIV-2-infected patients and in 9 out of 24 HIV-1-infected patients investigated.
NA, not applicable. Values are medians, with limits in brackets. Test cutoff value was used to calculate the median if levels were below cutoff. Significance in comparison to seronegative: *, P < 0.05; **, P < 0.01; ***, P < 0.0001. Significance in comparison to HIV-1: #, P < 0.05; ###, P < 0.0001.
TABLE 2.
Characterization of ART-treated HIV-2+ cohortsa
| Parameter | ART HIV-2 aviremic | ART HIV-2 viremic | ART HIV-2 (all) |
|---|---|---|---|
| No. (male/female) | 11 (5/6) | 5 (3/2) | 16 (8/8) |
| Median age (range) (yr) | 54 (34-62) | 53 (31-62) | 54 (31-62) |
| Current ART regimen (no. of patients) | |||
| 2 NRTI | 3 | 3 | |
| 2 NRTI + 1 PI | 6 | 4 | 10 |
| 3 NRTI | 2 | 1 | 3 |
| Length of follow-up (range) (mo) | |||
| Under ART | 28 (1-109) | 66 (6-120) | 58 (1-120) |
| After HIV diagnosis | 65 (8-242) | 177 (29-220) | 97 (8-242) |
| CD4 T cells/μl (range) | 340 (70-554) | 163 (84-719) | 302 (70-719) |
| % CD4 T cells (range) | 22 (5-36) | 10 (4-23) | 20 (4-36) |
| CD8 T cells/μl (range) | 825 (244-1,419) | 912 (447-1,532) | 887 (244-1,532) |
| % CD8 T cells (range) | 50 (26-64) | 53 (49-65) | 51 (26-65) |
| Viremia (RNA copies/ml) (range) | 200 | 4172 (499-34,314) | 200 (200-34,314) |
| Proviral DNA (copies/106 PBMC) (range)b | 108 (5-346) | 5 (5-726) | 57 (5-726.0) |
Values are medians, with limits in brackets. Test cutoff value was used to calculate the median if levels were below cutoff. NRTI, nucleoside RT inhibitor; PI, protease inhibitor.
Proviral DNA was below 5 copies/106 PBMC (cutoff) in 5 out of 11 aviremic and in 3 out of 5 viremic treated HIV-2 patients.
Cell isolation and flow cytometry.
Peripheral blood mononuclear cells (PBMC) were isolated immediately after blood collection and characterized by flow cytometry as previously described (9).
DNA and mRNA extraction.
DNA was extracted from 5 × 106 PBMC using the QIAamp DNA minikit. For mRNA extraction, 5 × 106 PBMC were lysed (RLT), homogenized (QIAshredder columns), extracted (Oligotex mRNA direct minikit) (all from Qiagen, Valencia, CA), treated with DNase (DNA-free kit; Ambion, Austin, TX), and immediately converted to cDNA. Samples were quantified using a NanoDrop ND-1000 spectrophotometer (NanoDrop technologies, Wilmington, DE).
Viremia and proviral DNA.
HIV-1 viremia was quantified by a reverse transcriptase PCR (RT-PCR) assay with a detection threshold of 40 RNA copies/ml (Roche, Basel, Switzerland). HIV-2 viremia was quantified using a previously described in-house-developed assay with a detection threshold of 200 RNA copies/ml (54). Quantification of viremia in ART-treated individuals with levels below 200 RNA copies/ml was repeated using a more sensitive real-time RT-PCR assay with a detection threshold of 40 RNA copies/ml as described previously (20). HIV-1 and HIV-2 total viral DNA (integrated and nonintegrated viral DNA species) was quantified using real-time PCR assays that amplify highly conserved regions in HIV-1 and HIV-2 gag with a detection range of 7 orders of magnitude and a sensitivity of 5 copies as we have previously described (52). Test cutoff values were used to calculate the median and the correlations with other parameters when levels were below the cutoff.
gag and tat mRNA.
Eighty ng of mRNA was reverse transcribed using the Superscript II reverse transcriptase kit (Invitrogen, Carlsbad, CA), and 250 nM random hexamers was quantified in duplicate using cDNA (1 μg) in a PCR mixture (50 μl) containing 25 μl Platinum quantitative PCR SuperMix-UDG, 1 μl ROX reference dye (50×), 5 mM MgCl2 (all from Invitrogen), and variable concentrations of the following primers and probes: HIV-1 gag F2 (10 pmol/μl), 5′-GGGAGAATTAGATCGATGGGAAA-3′; HIV-1 gag R1 (10 pmol/μl), 5′-GCTCCCTGCTTGCCCATA-3′; HIV-1 gag probe, 5′-6-carboxyfluorescein (FAM)-CCCTGGCCTTAACCGAATT-minor groove binder (MGB)-3′; HIV-2 gag F2 (10 pmol/μl), 5′-CGCGAGAAACTCCGTCTTG-3′; HIV-2 gag R2 (10 pmol/μl), 5′-CACACAATATGTTTTAGCCTGTACTTTTT; and probe HIV-2-gag, 5′-FAM-CCGGGCCGTAACCT-MGB-3′. tat multiply spliced (MS) mRNA expression was quantified using the following primers and probes: HIV-1 tat F2.3 (6 pmol/μl), 5′-GACGAAGAGCTCCTCAAGACA-3′; HIV-1 tat R2.4 (6 pmol/μl), 5′-GAGACAGAGACAGATCCGGTC-3′; HIV-1 tat probe, 5′-FAM-TCTCTATCAAAGCAACCCGC-MGB-3′; HIV-2 tat F3.5 (10 pmol/μl), 5′-AGGGGCTCGGGATATGTT-3′; HIV-2 tat R3.1 (10 pmol/μl), 5′-TCTGTATCCACCGTCGTTTC-3′; and HIV-2 tat probe, 5′-FAM-TGCATCAGACAAATC-MGB-3′, in an ABI Prism 7000 SDS (Applied Biosystems, Foster City, CA). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control (FAM/MGB probe, nonprimer limited; Applied Biosystems; standard curve with cDNA generated from pooled PBMC from 3 seronegatives). Standard curves for tat were generated using amplifications of synthetic oligonucleotides (GenScript Corporation, Piscataway, NJ) corresponding to 200-bp fragments of HIV-1 and HIV-2 tat transcripts, and those for gag were as described previously (52). The median values and the correlations with other parameters were calculated using the tat- and gag-relative quantification values of patients presenting detectable levels of the transcripts.
HIV-2 sequencing.
The in-house methodology was used to sequence a 1,280-bp HIV-2 pol gene fragment, including the entire protease and part of the reverse transcriptase from plasma samples. Viral RNA was retrotranscribed and amplified using the Access RT-PCR core reagents kit (Promega, Madison, WI) and outer primers JA218 ([+1859] 5′-GAA AGA AGC CCC GCA ACT TCC-3′) and JA221 ([−3258] 5′-GCT CTG CTT CTG CTA ATT CTG TCC A-3′) as described previously (54). A nested PCR was performed; a second amplification was carried out using AmpliTaq Gold PCR master mix and the inner primers JA219 [(+1898) 5′-AGG GGC T(A/G)A CAC CAA CAG CAC-3′] and JA220MOD [(−3178) 5′-GTC TTT ATI CCT GGG TAG AI(T/G) TGT G-3′]. Cycle sequencing was accomplished with the BigDye Terminator version 3.1 (v3.1) cycle sequencing kit according to the manufacturer's recommendations and using four sequencing primers: JA219, JA220 [(−3178) 5′-GTC TTT AT(T/C) CCT GGG TAG ATT TGT G-3′], JA222 ([+2525]) 5′-ACC TCC AAC TAA TCC TTA TAA TAC C-3′), and JA223 ([−2625] 5′-ACT GAA TTT CTG TGA AAT CTT GAG T-3′). Purified products were run on an ABI Prism 3100 genetic analyzer according to the same protocol but were adjusted to HIV-2-specific settings; nucleotide sequences were analyzed with SeqScape software version 2.5 (all from Applied Biosystems) by alignment with the ROD HIV-2 reference strain (GenBank accession number M15390).
Statistical analysis.
Statistical analysis was performed using GraphPad Prism version 5.00 (GraphPad Software, Inc., SD) using the Mann-Whitney test, Spearman's coefficient, and Fisher's exact test. P values of <0.05 were considered significant. The test cutoff value was used to calculate the median if levels were below the cutoff.
RESULTS
Ongoing viral replication in untreated HIV-2 infection.
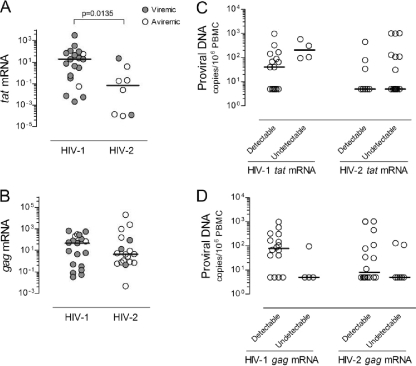
In order to investigate the degree of ongoing viral replication during the natural history of HIV-2 and HIV-1 infections, we compared untreated HIV-2 and HIV-1 cohorts matched for the degree of CD4 T cell depletion (Table 1), although length of infection was likely greater in HIV-2+ than in HIV-1+ individuals. As expected, viremia was significantly lower in HIV-2 than in HIV-1 infection (3, 5, 46, 50, 54). Twenty-two out of 29 HIV-2+ patients had levels below the test cutoff (aviremic), and the highest viremia was 2.6 × 104 RNA copies/ml (Table 1). Conversely, the proviral DNA loads, as previously reported, were similar (6, 24, 45, 52), suggesting comparable numbers of infected cells in these two groups despite their distinct viremias (Table 1). Of note, although no significant correlation was found between proviral DNA and viremia in HIV-2 infection, significantly higher proviral levels were found in viremic than in “aviremic” individuals (P = 0.0396).
tat mRNA, a multiply spliced (MS) HIV transcript, is thought to be expressed mainly in recently infected cells and/or cells actively producing virus (31, 32, 38, 53). We found a significantly higher number of individuals with undetectable tat mRNA in the HIV-2 than in the HIV-1 cohort (P = 0.0014; Fisher's exact test). Moreover, HIV-2+ patients with detectable tat mRNA featured significantly lower levels than their HIV-1+ counterparts (Fig. 1A), and a significant correlation with viremia was observed only with the HIV-1 cohort (r = 0.4791, P = 0.0443, n = 19 for the HIV-1 cohort; r = 0.0312, P = 0.9349, n = 8 for the HIV-2 cohort). Although no correlation with CD4 T cell levels were observed (Table 3), individuals with low CD4 T cell counts tended to have higher levels of tat mRNA (see Fig. 1 posted at http://www.imm.fm.ul.pt/dl/SoaresEtAlSupplementalFigure1AndLegend.pdf).
FIG. 1.
tat and gag mRNA expression and proviral DNA levels in untreated HIV-2 and HIV-1 infections. Graphs show the expression of tat (A) and gag (B) (as arbitrary units) relative to GAPDH in those individuals with detectable levels of these transcripts, from a total of 25 HIV-2+ and 23 HIV-1+ individuals evaluated. Open symbols refer to patients with viremia below the cutoff of the test, and closed symbols to patients with detectable viremia. Proviral DNA levels were compared for untreated HIV-2+ (25 of 25) and HIV-1+ (20 of 23) individuals with detectable and undetectable tat (C) and gag (D) mRNA transcripts. Bars represent medians.
TABLE 3.
Correlations between virological parameters and levels of CD4 T cells and immune activation in untreated HIV-2 and HIV-1 infections
| Viremia, DNA, or transcript | Virus type | r; Pa |
|||
|---|---|---|---|---|---|
| % CD4 T cells | CD4 T cells/μl | % HLA-DR+ in CD4 T cells | % HLA-DR+ 38+ in CD8 T cells | ||
| Viremia | HIV-2 | −0.3097; 0.1021 | −0.3943; 0.0343 | 0.5186; 0.0039 | 0.5288; 0.0032 |
| HIV-1 | −0.6120; 0.0009 | −0.5810; 0.0019 | 0.4731; 0.0146 | 0.7140; <0.0001 | |
| Proviral DNA | HIV-2 | 0.0313; 0.8719 | 0.1555; 0.4207 | 0.2281; 0.2339 | 0.1029; 0.5952 |
| HIV-1 | 0.0652; 0.7621 | −0.1019; 0.6358 | 0.0005; 0.9983 | 0.1590; 0.4580 | |
| tat mRNA | HIV-2 | −0.5952; 0.1323 | −0.5238; 0.1966 | 0.6190; 0.1017 | 0.6667; 0.0831 |
| HIV-1 | −0.3737; 0.1150 | −0.3193; 0.1827 | 0.2825; 0.2413 | 0.5404; 0.0169 | |
| gag mRNA | HIV-2 | −0.5728; 0.0130 | −0.3870; 0.1126 | 0.4985; 0.0353 | 0.3189; 0.1971 |
| HIV-1 | 0.2281; 0.3477 | 0.1869; 0.4435 | −0.4018; 0.0882 | −0.4193; 0.0739 | |
Test cutoff values were used to calculate the correlations with viremia or proviral DNA, if levels were below the cutoff. Correlations with viral mRNA were calculated using the tat and gag relative quantification values of patients presenting detectable levels of the transcripts (8 and 18 in HIV-2, respectively, and 19 for both tat and gag in HIV-1). Statistically significant correlations are represented in bold.
Conversely, gag mRNA, an unspliced (US) HIV transcript, was similarly expressed in the HIV-1 and HIV-2 cohorts (Fig. 1B), which included an equal number of individuals with undetectable expression levels. Of note, no correlation with viremia was found for either infection.
Furthermore, on comparing viremic and aviremic HIV-2+ individuals, we observed no significant differences between the number of patients with detectable tat and gag mRNA transcripts, and we found similar levels of expression of tat and gag mRNA (Fig. 1A and B), although these results should be interpreted cautiously, given the small number of individuals assessed.
A low ratio of gag to tat mRNA levels has been suggested as a marker of active viral transcription and of HIV-1 disease progression (22, 37, 49). We found a significantly higher gag-to-tat ratio in HIV-2+ than in HIV-1+ patients (P = 0.0118), although no association with viremia was found in either cohort.
Of note, neither tat nor gag mRNA expression levels nor the gag-to-tat ratio significantly correlated with the proviral DNA load in either HIV cohort. Moreover, when we subdivided the HIV-1 and HIV-2 cohorts on the basis of detectable and undetectable viral mRNA, no differences in proviral DNA levels were observed (Fig. 1C and D). Thus, no direct relationship appears to exist between the number of infected cells, as estimated by proviral DNA, and the levels of tat and gag transcription in PBMC during untreated HIV-2 and HIV-1 infections, suggesting a significant contribution of archived viral DNA.
Overall, HIV-2+ individuals exhibited reduced levels of tat and similar levels of gag transcripts compared to their HIV-1+ counterparts, translating into a higher gag-to-tat mRNA ratio in the former.
Relationship of plasma and cell-associated viral load with CD4 T cell depletion and T cell activation in HIV-2 infection.
We have previously shown that T cell activation markers were similarly upregulated in HIV-1 and HIV-2 infections when patients were matched for CD4 T cell levels, suggesting that CD4 T cell depletion is more directly linked to immune activation than to viral load (28, 55). Here, we assessed the relationship between cell-associated viral mRNA and DNA and the hyperimmune activation observed for HIV-2 and HIV-1 infections.
With respect to gag mRNA expression, we found a direct correlation with the levels of HLA-DR expression (Table 3) and HLA-DR and CD38 coexpression (r = 0.5150; P = 0.0287) within CD4 T cells in HIV-2+ individuals; this correlation did not reach statistical significance in HIV-1+ individuals. No correlations were found with CD8 T cell activation levels (Table 3).
Conversely, we observed that the levels of tat mRNA in HIV-1+ individuals directly correlated with CD8 T cell activation levels, measured either as the proportion of HLA-DR+ CD38+ (Table 3) or CD38+ cells (r = 0.6196; P = 0.0047), as well as CD38 mean fluorescence intensity (MFI; r = 0.7386; P = 0.0003). Clear trends for an association between tat mRNA levels and CD8 T cell activation were also observed with the HIV-2 cohort, but no significant correlations were found with CD4 T cell activation in either infection (Table 3).
Of note, proviral DNA levels did not directly correlate with either CD4 or CD8 T cell activation in HIV-1+ and HIV-2+ individuals (Table 3), further suggesting that a significant component comprises archived quiescent virus.
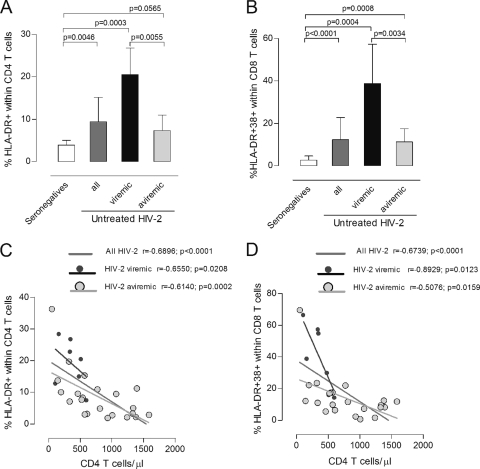
In spite of the narrow range of HIV-2 viremia (undetectable to 26,263 copies/ml), significant positive correlations were found between viremia and several T cell activation markers (Table 3) (percentage of HLA-DR+ CD38+ within CD4 T cells: r = 0.5685, P = 0.0013; percentage and MFI of CD38 within CD8 T cells: r = 0.6196, P = 0.0047, and r = 0.5330, P = 0.0029, respectively). In order to further assess the relative contribution of viremia, we compared viremic and aviremic HIV-2+ individuals (Table 1). Although the former showed significantly higher CD4 and CD8 T cell activation than the latter (Fig. 2A and B), both viremic and aviremic HIV-2+ individuals had significantly higher levels of T cell activation than seronegatives did (Fig. 2A and B) and exhibited strong negative correlations between CD4 T cell numbers and CD4 and CD8 T cell activation levels (Fig. 2C and D).
FIG. 2.
Relationship between viremia, absolute CD4 T cell numbers, and T cell activation in untreated HIV-2 infection. Proportion of CD4 T cells expressing HLA-DR (A) and CD8 T cells coexpressing HLA-DR and CD38 (B) in seronegative individuals (white bars) and untreated HIV-2 patients (dark gray bars). HIV-2 patients were further stratified into viremic (black bars) and aviremic groups (light gray bars). Bars represent median ± interquartile range. Correlations between CD4 T cell depletion and proportion of cells expressing HLA-DR within the CD4 T cell subset (C) and proportion of CD8 T cells coexpressing HLA-DR and CD38 (D).
Overall, our data not only demonstrated that T cell activation was strongly associated with CD4 T cell depletion, both in viremic and aviremic HIV-2+ individuals, but also supported a contribution, even at low levels, of circulating virus to both CD4 and CD8 T cell activation. Moreover, we showed that gag mRNA was directly related to CD4 T cell activation and tat mRNA to CD8 T cell activation, suggesting an overall impact of viral transcripts upon T cell activation.
Cell-associated viral mRNA and DNA in HIV-2+ patients under ART.
In order to further dissect the impact of cell-associated viral DNA and RNA upon HIV-2 immunopathogenesis, we assessed these parameters in patients receiving ART (Table 2). This cohort exhibited significantly lower CD4 T cell counts than untreated HIV-2+ individuals did (P = 0.0046), in agreement with previous reports showing a limited CD4 T cell recovery in ART-treated HIV-2 infection (1, 19, 29, 40, 51, 56). Viremia levels were similar in the treated and untreated HIV-2 cohorts, due to the low-level viremia detected in some of the ART-treated HIV-2+ patients (Table 2).
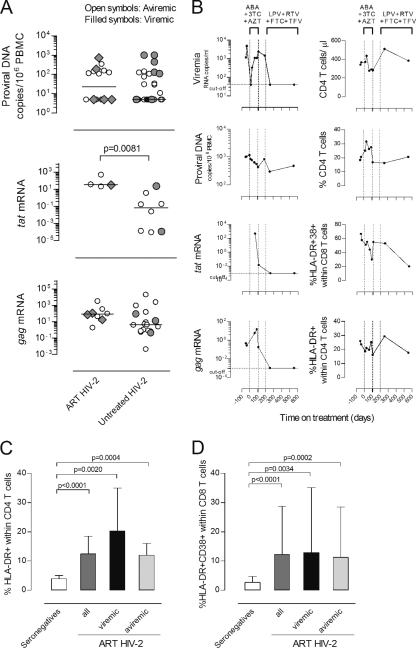
Of note, in contrast with the progressive decline in proviral DNA levels usually observed with ART-treated HIV-1+ patients (23, 58), proviral DNA did not significantly differ between untreated and treated HIV-2+ patients (Fig. 3A), despite the prolonged treatment. Moreover, treated and untreated HIV-2 patients featured similar levels of gag mRNA expression, whereas tat expression was significantly higher in the treated cohort, although the reduced number of individuals with detectable tat transcripts preclude a definitive interpretation (Fig. 3A). No significant differences were found between the number of patients with detectable tat and gag mRNA transcripts within treated and untreated groups. These results showed significant amounts of ongoing viral replication in ART-treated HIV-2 infection, suggesting a limited effectiveness of the ART regimens in HIV-2 infection, which justifies further exploration in large cohorts treated with optimized ART regimens.
FIG. 3.
Impact of ART on HIV-2 plasma and cell-associated viral load, CD4 T cell frequency, and T cell activation. (A) Levels of proviral DNA and tat and gag mRNA (expressed as arbitrary units) were compared in untreated and ART-treated HIV-2+ individuals. Open symbols represent patients with viremia below the cutoff of the test, and closed symbols patients with detectable viremia. Bars represent medians. (B) Longitudinal study of a representative ART-treated HIV-2+ patient. Day 0 was defined as the therapy initiation date. The vertical dashed lines indicate the periods under specific ART regimens (ABC, abacavir; 3TC, lamivudine; AZT, zidovudine; LPV, lopinavir; RTV, ritonavir; FTC, emtricitabine; TDF, tenofovir). Mutations in protease (PR) and reverse transcriptase (RT) were analyzed before therapy (day −37) and after the first ART regimen (day 108). Mutations [amino acid changes] found before therapy (day −37) were, in PR, Y14H, N40S, N41D, N68G, K69[K, R], and K70R and, in RT, D17[E, D], W24[G, W], K28R, C38[W, C], K43[K, R], K64R, L74V, K82R, D123G, P126Q, H162Y, V167I, K176S, H228D, W235[G, C, W], and Q245E. Mutations found after first therapy (day 108) were, in PR, Y14H, N40S, N41D, N68G, K69R, and K70R and, in RT, K28R, K43[K, R], K64R, W71R, L74I, K82R, T88[T, P], H96[H, P], G99[A, G], D123G, P126[Q, P], I159[I, L], H162Y, V167I, K176S, M184V, I187[I, L], and L209[P, L]. Proportion of CD4 T cells expressing HLA-DR (C) and CD8 T cells coexpressing HLA-DR and CD38 (D) in seronegative individuals (white bars) and ART-treated HIV-2+ individuals (dark gray bars). HIV-2+ patients were further stratified into viremic (black bars) and aviremic groups (light gray bars). Bars represent median ± interquartile range.
Additionally, neither the amount of gag and tat transcripts nor proviral DNA significantly differed between treated HIV-2 patients with detectable and undetectable viremia (Fig. 3A), emphasizing that the absence of detectable viremia is not a good surrogate marker for cell-associated viral mRNA and/or DNA during HIV-2 infection.
A rapid emergence of mutations potentially associated with drug resistance has been reported for HIV-2+ patients under ART (25, 29, 41). We confirmed the presence of mutations in the reverse transcriptase and protease in our ART-treated HIV-2 cohort (Table 4). These data further support the presence of significant amounts of ongoing replication in these ART-treated HIV-2+ patients, leading us to speculate about the potential contribution of ART-induced selective pressure upon the virus to the increased tat transcript levels (Fig. 3).
TABLE 4.
Virological parameters and CD4 T cell levels of HIV-2+ patients under ARTd
| Case no. | Current ART regimen | Follow-up after HIV diagnosis/under ART (mo)a | CD4 T pre-ART; post-ART (cells/μl)a | Mutations | Viremia (RNA copies/ml) | Proviral DNA (copies/106 PBMC) | tat mRNA (arbitrary units) | gag mRNA (arbitrary units) |
|---|---|---|---|---|---|---|---|---|
| 1 | d4T + 3TC + SQV/rb | 177/108 | 300; 719 | NA | 4,172 | 5 | Undet | 1.07 × 102 |
| 2 | AZT/3TC/ABC | 63/63 | 138; 554 | NA | <40 | 5 | Undet | 9.82 × 101 |
| 3 | AZT/3TC | 26/26 | 457; 535 | PR + RTc | <40 | 5 | 2.13 × 101 | Undet |
| 4 | AZT/3TCb | 206/64 | 500; 520 | NA | 44 | 158 | Undet | Undet |
| 5 | AZT/3TC + LPV/r | 126/26 | 125; 457 | NA | <40 | 114 | Undet | 3.56 × 102 |
| 6 | AZT/3TC + LPV/r | 38/28 | 164; 406 | NA | <40 | 108 | Undet | 4.46 × 101 |
| 7 | AZT/3TC/ABC | 242/61 | 287; 340 | PR + RTc | 191 | 5 | ND | ND |
| 8 | AZT/3TC + SQV/r | 8/5 | 222; 326 | NA | <40 | 145 | Undet | Undet |
| 9 | AZT/3TC + NFVb | 29/4 | 143; 277 | PR + RTc | 499 | 5 | 3.14 × 101 | 1.72 × 101 |
| 10 | AZT/3TC | 15/1 | 297; 273 | NA | <40 | 126 | Undet | Undet |
| 11 | AZT/3TC + IDV/r | 65/51 | 112; 251 | NA | <40 | 346 | 5.19 × 102 | 1.91 |
| 12 | AZT/3TC/ABC | 220/56 | 176; 163 | PR + RTc | 742 | 187 | Undet | 7.25 × 101 |
| 13 | AZT/3TC + IDVb | 218/6 | 121; 102 | PR + RTc | <40 | 5 | 3.73 × 101 | 3.30 × 103 |
| 14 | d4T + 3TC + SQV/rb | 177/120 | 82; 87 | NA | 34,314 | 726 | ND | ND |
| 15 | AZT/3TC + LPV/rb | 67/12 | 120; 84 | PR + RTc | 10,474 | 5 | Undet | Undet |
| 16 | ABC/FTC + LPV/rb | 216/7 | 128; 70 | NA | <40 | 5 | Undet | Undet |
ART refers to the current therapy.
Previous ART regimens included the following: case 1, AZT, ddI, ddC; case 4, ddC, d4T; case 9, ddI, EFV; case 13, d4T; case 14, AZT, ddI; case 15, ddI, d4T, NFV; case 16, AZT.
The following mutations (amino acid change) were found (those mutations previously associated with drug resistance are in bold). Case 3: for PR, P19S, N61D, E65K, L99F; for RT, V5L, K9R, I10V, M11T, E44[K, R, E, G], K68G, R72[K, R], K82R, D86E, Q91[Q, P], P126Q, H162Y, V167I, K173R, K176Q, I179T, E197G, R200K, Y227F, H228Q. Case 7: for PR, Y14[H, Y], N40S, N61D, E65[K, R], K70R; for RT, M11T, E44[E, D], K82R, P126Q, H162Y, V167I, K176Q, M184V, D218[E, D], E219D, D224[N, D], P226[P, S], H228Q, W239[G, W], Q245H, L246W, Q248L, E250D, I251T, K259N, N265I, W266G. Case 9: for PR, E21D, N40S, T56[T, A], V62[A, V], K70R, V71I, L99F; for RT, V5I, M11T, D17[E, D], P51S, D76V, F77L, R78G, E79K, P126Q, S134A, V167I, K176P, I180L, M184V, H228Q, E250G, I251V, K259N, N265I. Case 12: for PR, Y14H, N40S, N41[N, D], I46[I, V], N61[N, D], E65R, D79E; for RT, K9R, I10V, M11T, R22K, K35R, K40R, P51S, K70Q, D86E, R104[K, R], V111I, P126Q, S134A, V167I, K176P, I180[I, L], M184V, S215Y, F221L, D224N, H228Q. Case 13: PR, Y14H, N40S, E65R, K70R, F85L, I89V, L90 M; for RT, A3V, V5M, I10V, M11R, K43R, T53S, T58S, T60[T, P], T88[T, P], P126Q, A138T, V167I, A174T, K176P, I180L, M184V, H228Q, K243[K, R]. Case 15: for PR, K7R, V10I, V22I, V33I, E37D, N40S, I54 M, T56V, N68G, K70R, V71I, M76L, I82F, I89[I, V], T91A, G94[S, G]; for RT, P1[P, L], V5[I, V], I10[I, V], D17[N, D], E48[E, G], A62[A, V], K64[K, R], K65[K, R], A101P, P126Q, P126Q, V135A, Q151M, H162Y, V167I, K176P, I180L, M184V, I189L, V201A, F214L, D218[K, N, E, D], E219[E, D], H228K, V254F.
ND, not done; Undet, undetectable mRNA expression; NA, no amplification; ART, antiretroviral therapy; reverse transcriptase (RT) inhibitors: AZT, zidovudine; 3TC, lamivudine; ABC, abacavir; d4T, stavudine; ddC, zalcitabine; ddI, didanosine; EFV, efavirenz; FTC, emtricitabine; r, ritonavir. Protease (PR) inhibitors: LPV, lopinavir; IDV, indinavir; NFV, nelfinavir; SQV, saquinavir.
The selection of adequate ART regimens to treat HIV-2 infection is difficult given the lack of clinical trials. There are currently some phenotypic studies that allow the selection of better first-line therapies than those used a few years ago (16, 61), but the knowledge of HIV-2 resistance pathways is still incomplete. Longitudinal data of an individual HIV-2+ patient, in whom the initial ART regimen was associated with the rapid emergence of mutations and virological failure, illustrated that switching to another ART combination was associated with viremia suppression and a decline of cell-associated viral burden (Fig. 3B). This was supported by both a loss of measurable tat and gag transcripts and a decrease in the levels of proviral DNA, both of which are indicative of a successful virological response. Nevertheless, this patient's immunological response was limited, with only marginal CD4 T cell recovery and a partial decrease in both CD4 and CD8 T cell immune activation (Fig. 3B), suggesting that factors other than viral replication are contributing to the poor immunological reconstitution. Our findings support a role for persistent immune activation, given that, as shown in Fig. 3C and D, ART-treated HIV-2+ individuals exhibited significantly higher levels of T cell activation than seronegatives did, similar levels of CD4 and CD8 T cell activation in comparison with the untreated HIV-2 cohort, and significant negative correlations between absolute CD4 T cell numbers and the percentages of HLA-DR+ within CD4 T cells (r = −0.5147; P = 0.0413) and HLA-DR+ CD38+ within CD8 T cells (r = −0.5294; P = 0.0350).
Overall, significant amounts of ongoing viral replication were observed with HIV-2-infected patients under ART, highlighting the importance of additional studies of antiretroviral drug efficacy in HIV-2 infection. Furthermore, ART apparently failed to have a significant impact on T cell activation, even in patients with undetectable viremia, possibly contributing to their low CD4 T cell recovery.
DISCUSSION
Here we demonstrated that untreated HIV-1+ and HIV-2+ individuals with similar degrees of CD4 T cell depletion featured similar levels of gag mRNA transcripts, suggesting that significant viral transcription occurs in HIV-2 patients, despite the lower viremia. Conversely, we found decreased levels of tat mRNA in untreated HIV-2+ individuals. Given the previous reports demonstrating that HIV-1 tat mRNA transcripts accumulate and outnumber gag mRNA transcripts in recently infected cells (31, 32, 38, 53), our results provide evidence for a decreased rate of de novo cell infection in HIV-2 disease.
Proviral DNA levels have been used to estimate the extent of viral reservoirs (8, 10, 11, 18). We and others have shown that the levels of total proviral DNA are similar in the two HIV infections despite the reduced viremia observed with HIV-2+ individuals (6, 45, 52). This may result from a preferential contribution of latently infected, quiescent T cells to total HIV-2 proviral DNA. Alternatively, a significant amount of ongoing viral replication occurs, but it does not translate into plasma viral load. In this study, we showed that the levels of proviral DNA were not associated with viral transcription levels in HIV-1+ or in HIV-2+ individuals, bringing into question the reliability of proviral DNA levels as a marker of replicative activity in both infections.
Additionally, the impact of plasma and cell-associated viral load upon HIV-2-associated hyperimmune activation was investigated. Of note, significantly higher levels of CD4 and CD8 T cell activation were found in viremic individuals than in aviremic individuals, despite the small amount of circulating virus observed with viremic HIV-2+ patients. In agreement with a recent report on an African cohort (33), our data support a contribution of plasma viral load, even at low levels, to immune activation, which may be related to transmission of cell free viruses and/or immunological effects of viral proteins.
The direct association between gag mRNA and CD4 T cell activation that we observed with HIV-infected patients raises the possibility that ongoing viral replication significantly contributes to the maintenance of heightened T cell activation in HIV-2+ individuals in spite of the reduced viremia. In addition, the direct association found between tat mRNA levels and CD8 T cell activation, particularly in HIV-1+ individuals, suggests a specific role of this transactivator molecule and/or newly infected cells in driving CD8 T cell activation.
With respect to HIV-1 infection, those rare (<0.1%) individuals who are able to control viral replication in the absence of ART (27, 42, 44, 60) provide another valuable resource for the investigation of factors associated with viremia control. Various criteria have been used to define this population, including viremia ranging from undetectable (elite controllers) up to 2,000 RNA copies/ml (viremic controllers) (42, 60). Notably, the small cohort of HIV-1 controllers within our untreated HIV-1 cohort resembles the untreated HIV-2 cohort in terms of viremia, gag and tat mRNA expression, gag-to-tat ratio, and proviral DNA levels. The seven individuals with viremia of <2,000 RNA copies/ml tended to have lower levels of tat mRNA (14.59 ± 14.47 arbitrary units) than the other HIV-1+ patients did (262.0 ± 210.4 arbitrary units), though the levels did not reach statistical significance. The numbers of patients with detectable tat mRNA, gag mRNA, and proviral DNA were similar within these subgroups of the HIV-1 cohort, and no differences in gag mRNA levels or proviral DNA were observed, but the gag-to-tat ratio was higher in controllers (P = 0.0434). These data further support that viremia control is associated with reduced levels of tat transcripts.
Finally, we provide evidence of persistent HIV-2 replication during ART, based on proviral DNA and gag and tat mRNA levels, irrespective of detectable plasma viremia. We also demonstrated drug-related genetic evolution of HIV-2 reverse transcriptase and protease gene sequences. The high levels of tat mRNA observed with the ART-treated HIV-2 cohort suggested that the therapeutic regimens used were unable to reduce the rate of de novo cell infection. These data contrast with those for ART-treated HIV-1+ patients, in which virological response is usually associated with a sharp decline in MS mRNA (4, 21, 62) and in the proportion of MS mRNA relative to US mRNA in PBMC (57), as well as with a progressive decrease in proviral DNA (23, 58, 59) despite the low-level viremia that can frequently be detected using ultrasensitive assays (17, 43).
In agreement with previous reports (1, 19, 29, 40, 51), we documented poor CD4 T cell recovery in ART-treated HIV-2+ individuals, even in those with evidence of viral suppression. Our findings suggest that persistent hyperimmune activation may be a main determinant of this impaired immune reconstitution. The study of lymphoid tissue from HIV-2-infected patients will be instrumental in evaluating the degree of irreversible damage associated with long-term infection that may limit the potential for immune recovery and providing support for an early start of ART in this otherwise relatively benign disease.
In conclusion, we provide here evidence of ongoing viral replication in HIV-2 infection despite the low or undetectable viremia and of its association with CD4 and CD8 T cell activation, with the latter being more closely related to the levels of tat mRNA. Of particular note was the persistent viral replication in ART-treated HIV-2+ individuals. In light of these findings, particularly the apparent ease with which the virus mutated in treated HIV-2+ individuals to escape drug activity, there is an obvious need for large-scale drug trials for HIV-2 infection to determine the most appropriate drug regimen and also the benefit of an early initiation of therapy for this infection.
Acknowledgments
We thank Sara Sousa and Luis França from the Clínica Universitária de Doenças Infecciosas da Faculdade de Medicina da Universidade de Lisboa, as well as Luis Pinheiro from the Clínica Universitária de Medicina 2 from the Hospital Universitário de Santa Maria for collaboration in the collection of clinical data or patient follow-up and Nuno Taveira for helpful discussions.
This study was funded by grants from Fundação para a Ciência e a Tecnologia (FCT) and Programa Operacional Ciência e Inovação 2010 (POCI2010), as well as from Fundação Calouste Gulbenkian, to A.E.S. A.P.B. received a scholarship from GlaxoSmithKline, and R.S.S., R.T., R.B.F., and R.C. a scholarship from FCT.
All authors report no potential conflicts of interest.
Footnotes
Published ahead of print on 15 December 2010.
REFERENCES
- 1.Adje-Toure, C. A., et al. 2003. Antiretroviral therapy in HIV-2-infected patients: changes in plasma viral load, CD4+ cell counts, and drug resistance profiles of patients treated in Abidjan, Cote d'Ivoire. AIDS 17(Suppl. 3):S49-S54. [PubMed] [Google Scholar]
- 2.Adjorlolo-Johnson, G., et al. 1994. Prospective comparison of mother-to-child transmission of HIV-1 and HIV-2 in Abidjan, Ivory Coast. JAMA 272:462-466. [PubMed] [Google Scholar]
- 3.Andersson, S., et al. 2000. Plasma viral load in HIV-1 and HIV-2 singly and dually infected individuals in Guinea-Bissau, West Africa: significantly lower plasma virus set point in HIV-2 infection than in HIV-1 infection. Arch. Intern. Med. 160:3286-3293. [DOI] [PubMed] [Google Scholar]
- 4.Bagnarelli, P., et al. 1996. Dynamics and modulation of human immunodeficiency virus type 1 transcripts in vitro and in vivo. J. Virol. 70:7603-7613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berry, N., et al. 1998. Low peripheral blood viral HIV-2 RNA in individuals with high CD4 percentage differentiates HIV-2 from HIV-1 infection. J. Hum. Virol. 1:457-468. [PubMed] [Google Scholar]
- 6.Berry, N., et al. 1994. HIV type 2 proviral load measured by quantitative polymerase chain reaction correlates with CD4+ lymphopenia in HIV type 2-infected individuals. AIDS Res. Hum. Retroviruses 10:1031-1037. [DOI] [PubMed] [Google Scholar]
- 7.Brandin, E., et al. 2003. pol gene sequence variation in Swedish HIV-2 patients failing antiretroviral therapy. AIDS Res. Hum. Retroviruses 19:543-550. [DOI] [PubMed] [Google Scholar]
- 8.Brenchley, J. M., et al. 2004. T-cell subsets that harbor human immunodeficiency virus (HIV) in vivo: implications for HIV pathogenesis. J. Virol. 78:1160-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cavaleiro, R., et al. 2009. Major depletion of plasmacytoid dendritic cells in HIV-2 infection, an attenuated form of HIV disease. PLoS Pathog. 5:e1000667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chun, T. W., et al. 1997. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature 387:183-188. [DOI] [PubMed] [Google Scholar]
- 11.Chun, T. W., et al. 1997. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc. Natl. Acad. Sci. U. S. A. 94:13193-13197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clavel, F., et al. 1987. Human immunodeficiency virus type 2 infection associated with AIDS in West Africa. N. Engl. J. Med. 316:1180-1185. [DOI] [PubMed] [Google Scholar]
- 13.Colson, P., et al. 2005. Polymorphism and drug-selected mutations in the reverse transcriptase gene of HIV-2 from patients living in southeastern France. J. Med. Virol. 75:381-390. [DOI] [PubMed] [Google Scholar]
- 14.Colson, P., et al. 2004. Polymorphism and drug-selected mutations in the protease gene of human immunodeficiency virus type 2 from patients living in Southern France. J. Clin. Microbiol. 42:570-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Damond, F., et al. 2005. Polymorphism of the human immunodeficiency virus type 2 (HIV-2) protease gene and selection of drug resistance mutations in HIV-2-infected patients treated with protease inhibitors. J. Clin. Microbiol. 43:484-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Desbois, D., et al. 2008. In vitro phenotypic susceptibility of human immunodeficiency virus type 2 clinical isolates to protease inhibitors. Antimicrob. Agents Chemother. 52:1545-1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dinoso, J. B., et al. 2009. Treatment intensification does not reduce residual HIV-1 viremia in patients on highly active antiretroviral therapy. Proc. Natl. Acad. Sci. U. S. A. 106:9403-9408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Douek, D. C., et al. 2002. HIV preferentially infects HIV-specific CD4+ T cells. Nature 417:95-98. [DOI] [PubMed] [Google Scholar]
- 19.Drylewicz, J., et al. 2008. Comparison of viro-immunological marker changes between HIV-1 and HIV-2-infected patients in France. AIDS 22:457-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferns, R. B., and J. A. Garson. 2006. Development and evaluation of a real-time RT-PCR assay for quantification of cell-free human immunodeficiency virus type 2 using a Brome Mosaic Virus internal control. J. Virol. Methods 135:102-108. [DOI] [PubMed] [Google Scholar]
- 21.Fischer, M., et al. 2008. Biphasic decay kinetics suggest progressive slowing in turnover of latently HIV-1 infected cells during antiretroviral therapy. Retrovirology 5:107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Furtado, M. R., L. A. Kingsley, and S. M. Wolinsky. 1995. Changes in the viral mRNA expression pattern correlate with a rapid rate of CD4+ T-cell number decline in human immunodeficiency virus type 1-infected individuals. J. Virol. 69:2092-2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garrigue, I., et al. 2000. Cell-associated HIV-1-DNA quantitation after highly active antiretroviral therapy-treated primary infection in patients with persistently undetectable plasma HIV-1 RNA. AIDS 14:2851-2855. [DOI] [PubMed] [Google Scholar]
- 24.Gomes, P., et al. 1999. Quantitation of human immunodeficiency virus type 2 DNA in peripheral blood mononuclear cells by using a quantitative-competitive PCR assay. J. Clin. Microbiol. 37:453-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gottlieb, G. S., et al. 2009. Emergence of multiclass drug-resistance in HIV-2 in antiretroviral-treated individuals in Senegal: implications for HIV-2 treatment in resouce-limited West Africa. Clin. Infect. Dis. 48:476-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gottlieb, G. S., et al. 2006. Lower levels of HIV RNA in semen in HIV-2 compared with HIV-1 infection: implications for differences in transmission. AIDS 20:895-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grabar, S., et al. 2009. Prevalence and comparative characteristics of long-term nonprogressors and HIV controller patients in the French Hospital Database on HIV. AIDS 23:1163-1169. [DOI] [PubMed] [Google Scholar]
- 28.Grossman, Z., M. Meier-Schellersheim, A. E. Sousa, R. M. Victorino, and W. E. Paul. 2002. CD4+ T-cell depletion in HIV infection: are we closer to understanding the cause? Nat. Med. 8:319-323. [DOI] [PubMed] [Google Scholar]
- 29.Jallow, S., et al. 2009. Virological response to highly active antiretroviral therapy (HAART) in HIV-2 and HIV-1/HIV-2 dually infected patients in the Gambia and the emergence of drug-resistant variants. J. Clin. Microbiol. 47:2200-2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanki, P. J., et al. 1994. Slower heterosexual spread of HIV-2 than HIV-1. Lancet 343:943-946. [DOI] [PubMed] [Google Scholar]
- 31.Kim, S. Y., R. Byrn, J. Groopman, and D. Baltimore. 1989. Temporal aspects of DNA and RNA synthesis during human immunodeficiency virus infection: evidence for differential gene expression. J. Virol. 63:3708-3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klotman, M. E., et al. 1991. Kinetics of expression of multiply spliced RNA in early human immunodeficiency virus type 1 infection of lymphocytes and monocytes. Proc. Natl. Acad. Sci. U. S. A. 88:5011-5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leligdowicz, A., et al. 2010. Direct relationship between virus load and systemic immune activation in HIV-2 infection. J. Infect. Dis. 201:114-122. [DOI] [PubMed] [Google Scholar]
- 34.MacNeil, A., et al. 2007. Direct evidence of lower viral replication rates in vivo in human immunodeficiency virus type 2 (HIV-2) infection than in HIV-1 infection. J. Virol. 81:5325-5330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marlink, R., et al. 1994. Reduced rate of disease development after HIV-2 infection as compared to HIV-1. Science 265:1587-1590. [DOI] [PubMed] [Google Scholar]
- 36.Matheron, S., et al. 2003. Factors associated with clinical progression in HIV-2 infected-patients: the French ANRS cohort. AIDS 17:2593-2601. [DOI] [PubMed] [Google Scholar]
- 37.Michael, N. L., et al. 1995. Human immunodeficiency virus type 1 cellular RNA load and splicing patterns predict disease progression in a longitudinally studied cohort. J. Virol. 69:1868-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Michael, N. L., et al. 1991. Induction of human immunodeficiency virus type 1 expression in chronically infected cells is associated primarily with a shift in RNA splicing patterns. J. Virol. 65:1291-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Michael, N. L., M. Vahey, D. S. Burke, and R. R. Redfield. 1992. Viral DNA and mRNA expression correlate with the stage of human immunodeficiency virus (HIV) type 1 infection in humans: evidence for viral replication in all stages of HIV disease. J. Virol. 66:310-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mullins, C., et al. 2004. Highly active antiretroviral therapy and viral response in HIV type 2 infection. Clin. Infect. Dis. 38:1771-1779. [DOI] [PubMed] [Google Scholar]
- 41.Ntemgwa, M. L., T. d'Aquin Toni, B. G. Brenner, R. J. Camacho, and M. A. Wainberg. 2009. Antiretroviral drug resistance in human immunodeficiency virus type 2. Antimicrob. Agents Chemother. 53:3611-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Okulicz, J. F., et al. 2009. Clinical outcomes of elite controllers, viremic controllers, and long-term nonprogressors in the US Department of Defense HIV natural history study. J. Infect. Dis. 200:1714-1723. [DOI] [PubMed] [Google Scholar]
- 43.Palmer, S., et al. 2008. Low-level viremia persists for at least 7 years in patients on suppressive antiretroviral therapy. Proc. Natl. Acad. Sci. U. S. A. 105:3879-3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pereyra, F., et al. 2009. Persistent low-level viremia in HIV-1 elite controllers and relationship to immunologic parameters. J. Infect. Dis. 200:984-990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Popper, S. J., et al. 2000. Low plasma human immunodeficiency virus type 2 viral load is independent of proviral load: low virus production in vivo. J. Virol. 74:1554-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Popper, S. J., et al. 1999. Lower human immunodeficiency virus (HIV) type 2 viral load reflects the difference in pathogenicity of HIV-1 and HIV-2. J. Infect. Dis. 180:1116-1121. [DOI] [PubMed] [Google Scholar]
- 47.Rodés, B., et al. 2000. Emergence of drug resistance mutations in human immunodeficiency virus type 2-infected subjects undergoing antiretroviral therapy. J. Clin. Microbiol. 38:1370-1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ruelle, J., et al. 2008. Transmitted drug resistance, selection of resistance mutations and moderate antiretroviral efficacy in HIV-2: analysis of the HIV-2 Belgium and Luxembourg database. BMC Infect. Dis. 8:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Seshamma, T., O. Bagasra, D. Trono, D. Baltimore, and R. J. Pomerantz. 1992. Blocked early-stage latency in the peripheral blood cells of certain individuals infected with human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. U. S. A. 89:10663-10667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Simon, F., et al. 1993. Cellular and plasma viral load in patients infected with HIV-2. AIDS 7:1411-1417. [DOI] [PubMed] [Google Scholar]
- 51.Smith, N. A., et al. 2001. Antiretroviral therapy for HIV-2 infected patients. J. Infect. 42:126-133. [DOI] [PubMed] [Google Scholar]
- 52.Soares, R., et al. 2006. Increased frequency of circulating CCR5+ CD4+ T cells in human immunodeficiency virus type 2 infection. J. Virol. 80:12425-12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sonza, S., et al. 2002. Selectively reduced tat mRNA heralds the decline in productive human immunodeficiency virus type 1 infection in monocyte-derived macrophages. J. Virol. 76:12611-12621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Soriano, V., et al. 2000. Human immunodeficiency virus type 2 (HIV-2) in Portugal: clinical spectrum, circulating subtypes, virus isolation, and plasma viral load. J. Med. Virol. 61:111-116. [PubMed] [Google Scholar]
- 55.Sousa, A. E., J. Carneiro, M. Meier-Schellersheim, Z. Grossman, and R. M. Victorino. 2002. CD4 T cell depletion is linked directly to immune activation in the pathogenesis of HIV-1 and HIV-2 but only indirectly to the viral load. J. Immunol. 169:3400-3406. [DOI] [PubMed] [Google Scholar]
- 56.van der Ende, M. E., et al. 2003. Clinical, immunological and virological response to different antiretroviral regimens in a cohort of HIV-2-infected patients. AIDS 17(Suppl. 3):S55-S61. [DOI] [PubMed] [Google Scholar]
- 57.Vesanen, M., M. Markowitz, Y. Cao, D. D. Ho, and K. Saksela. 1997. Human immunodeficiency virus type-1 mRNA splicing pattern in infected persons is determined by the proportion of newly infected cells. Virology 236:104-109. [DOI] [PubMed] [Google Scholar]
- 58.Viard, J. P., et al. 2004. Impact of 5 years of maximally successful highly active antiretroviral therapy on CD4 cell count and HIV-1 DNA level. AIDS 18:45-49. [DOI] [PubMed] [Google Scholar]
- 59.Vitone, F., D. Gibellini, P. Schiavone, and M. C. Re. 2005. Quantitative DNA proviral detection in HIV-1 patients treated with antiretroviral therapy. J. Clin. Virol. 33:194-200. [DOI] [PubMed] [Google Scholar]
- 60.Walker, B. D. 2007. Elite control of HIV infection: implications for vaccines and treatment. Top. HIV Med. 15:134-136. [PubMed] [Google Scholar]
- 61.Witvrouw, M., et al. 2004. Susceptibility of HIV-2, SIV and SHIV to various anti-HIV-1 compounds: implications for treatment and postexposure prophylaxis. Antivir. Ther. 9:57-65. [PubMed] [Google Scholar]
- 62.Zanchetta, M., et al. 2006. Long-term decay of the HIV-1 reservoir in HIV-1-infected children treated with highly active antiretroviral therapy. J. Infect. Dis. 193:1718-1727. [DOI] [PubMed] [Google Scholar]