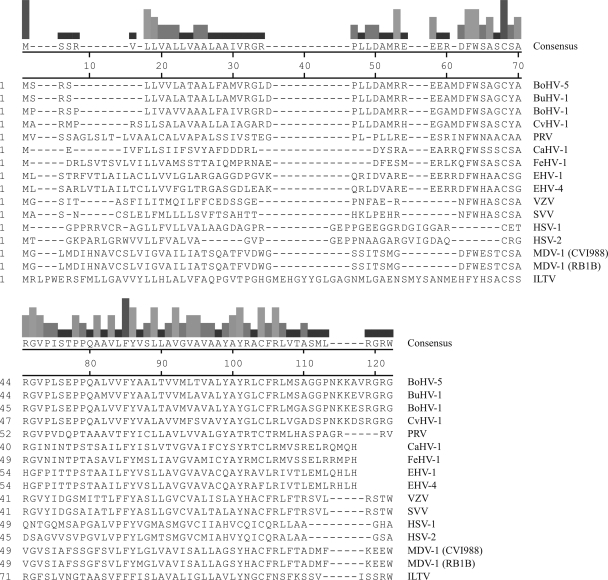
FIG. 1.
Alignment of the amino acid sequences of a selection of alphaherpesvirus UL49.5 proteins. The amino acid sequence alignment of UL49.5 homologs was performed by using CLUSTAL V of the MegAlign software from DNAStar. BoHV-5 UL49.5, bovine herpesvirus 5 (accession number NP_954898); BuHV-1 UL49.5, bubaline herpesvirus 1 (F. A. M. Rijsewijk, unpublished data); BoHV-1 UL49.5, bovine herpesvirus 1 (NP_045309); CvHV-1 UL49.5, cervid herpesvirus 1 (F. A. M. Rijsewijk, unpublished data); PRV/SuHV-1 UL49.5, pseudorabies virus (YP_068325); CaHV-1 UL49.5, canid herpesvirus 1 (patent EPO910406); FeHV-1 UL49.5, felid herpesvirus 1 (YP_003331529); EHV-1 UL49.5, equid herpesvirus 1 (YP_053055); EHV-4 UL49.5, equid herpesvirus 4 (NP_045227); VZV/HHV3 UL49.5, varicella-zoster virus (YP_068406); SVV UL49.5, simian varicella virus (NP_077423); HSV-1 UL49.5, herpes simplex virus 1 (NP_044652); HSV-2 UL49.5, herpes simplex virus 2 (NP_044520); MDV-1/GaHV-2 UL49.5, Marek's disease virus 1 strain CVI988 (ABF72292.1); MDV-1 strain RB1B (YP_001033979.1); ILTV/GaHV-1 UL49.5, infectious laryngotracheitis virus (YP_182341). The bars at the top of figure are proportional in height to the degree of homology of amino acid conservation among the different viral proteins.