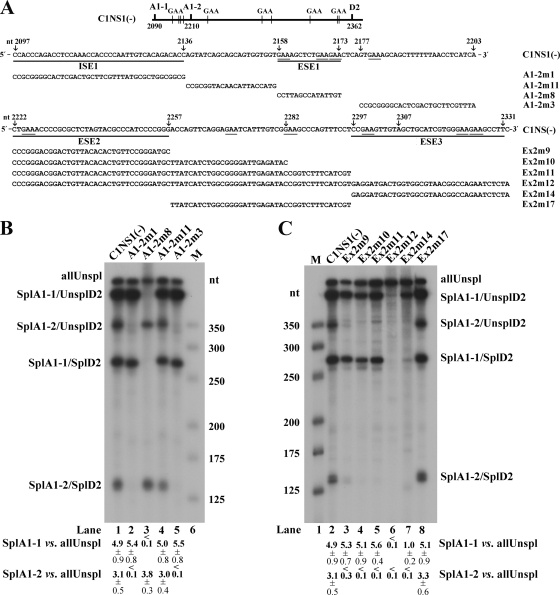
FIG. 2.
Exonic splicing enhancers (ESEs) in the central exon of the B19V genome. (A) Top, the B19V central exon (exon 2) is depicted, with the locations of splice sites and GAA motifs indicated. Middle, exon 2 sequence between the A1-1 and A1-2 sites (nt 2097 to 2203), with mutations in this region indicated (arrow plus nucleotide number) above the sequence, the identified ISE1 and ESE1 underlined, and mutant constructs tested in RNase protection assays identified and also shown below, in panel B. Bottom, exon 2 sequence between the A1-2 and D2 sites (nt 2222 to 2331), with mutations in this region indicated (arrow plus nucleotide number) above the sequence, identified ESE2 and ESE3 underlined, and mutant constructs tested in RNase protection assays identified and also shown below, in panel C. (B and C) RNase protection assays assessing the enhancer activities of the presence of ESE1 and ISE1 (B) and ESE2 and ESE3 (C). For these RNase protection assays, the C1NS1(−) plasmid and its mutation-containing derivatives (shown in panel A) were transfected into COS-7 cells. At 48 h posttransfection, total RNA was isolated and protected using probe 11. Transfections, RNA isolation, and RNase protection assays were performed as described previously (6, 11, 18). The protected bands are identified and labeled as the products described in the legend to Fig. 1. The size maker was made as previously described (16). A representative protection assay from at least three independent experiments is shown. Ratios of SplA1-1 (SplA1-1/UnsplD2 + SplA1-1/SplD2) to allUnspl and of SplA1-2 (SplA1-2/UnsplD2 + SplA1-2/SplD2) to allUnspl are shown, respectively, for each protection assay; some are given as averages and standard deviations.