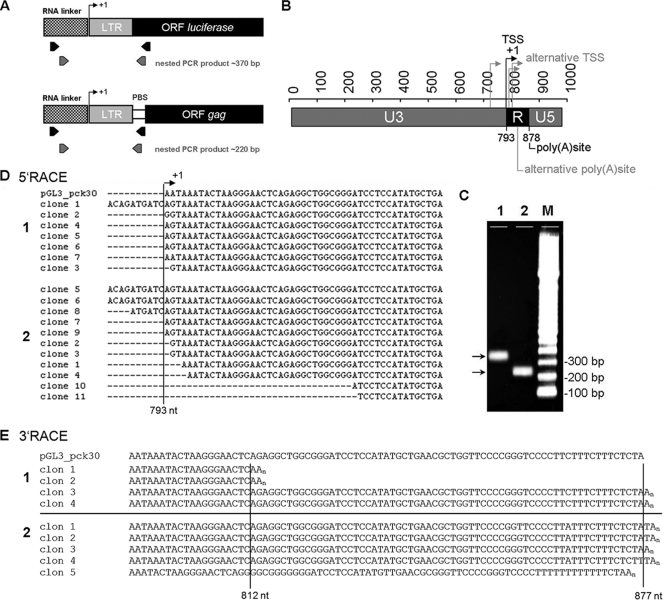
FIG. 4.
Definition of HERV-K LTR structure by 5′- and 3′-RACE. (A) Locations of the PCR primers used for amplification in 5′-RACE experiments. For endogenous transcripts, the reverse primer binds to the HERV-K PBS, whereas for amplification of cDNAs from pGL3_pck30 transfectants, the reverse primer binds within the luciferase gene. (B) Schematic presentation of redefined U3, R, and U5 regions. +1, major TSS at nt 793 in the 5′-LTR. The transcription termination site in the 3′-LTR is also plotted. Alternative TSS and termination sites are indicated in a light shade. (C) 5′-RACE nested PCR products of 370 bp for transcripts from the transfected plasmid pGL3_pck30 (lane 1) and of 220 bp for endogenous HERV-K transcripts (lane 2). (D) Sequences of clones from endogenous transcripts of GH (1) and Mel-C9 (2) cells in comparison to the 5′-RACE product obtained from the transfected pGL3_pck30 plasmid. The first bases transcribed from the pGL3_pck30 plasmid are AAT; transcription of endogenous proviruses started with AGT. This difference resulted from the LTRpck30 construct which combined the U3 region of the HERV-K 108 3′-LTR with the R/U5 region of its 5′-LTR. (E) 3′-RACE clones from Mel-C9 (1) and GH (2) cells, with the major polyadenylation site at nt 877 and a second polyadenylation site at nt 812 compared to the sequence of the pGL3_pck30 plasmid.