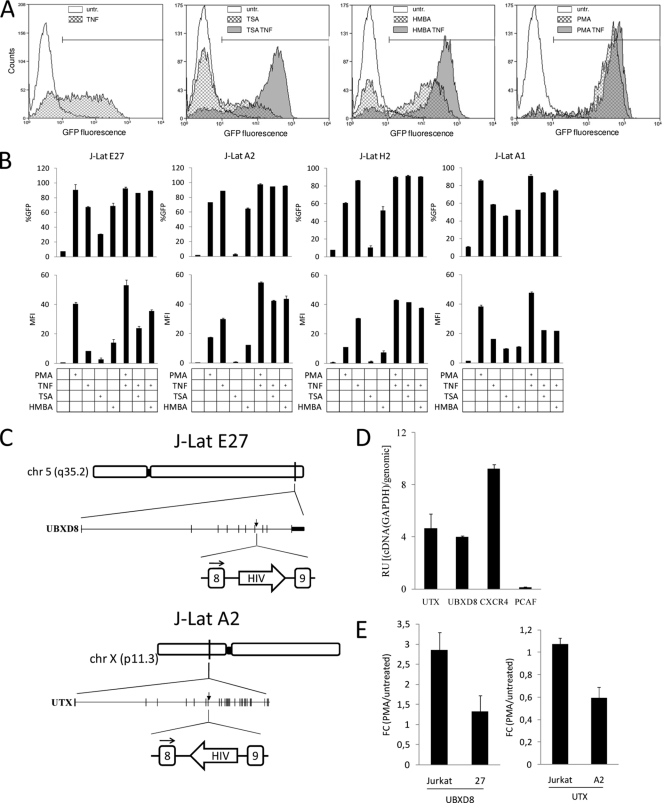
FIG. 1.
Characterization of latent HIV integration into introns of active genes. (A and B) Reactivation of Jurkat clones containing latent HIV minigenome integrations in different genome environments. Cells of clones J-Lat E27, A2, H2, and A1 were incubated with PMA (10 nM), TNF-α (10 ng/ml), TSA (400 nM), or HMBA (10 mM) for 16 h, and HIV-GFP reactivation was followed by fluorescence-activated cell sorting. Data are expressed as percentage of GFP-positive cells (%GFP) or mean fluorescence intensity (MFI). Values represent the mean and range of a representative experiment performed in duplicate. A representative fluorescence-activated cell sorting profile and gating on J-Lat E27 are shown in panel A. (C) Intronic integration of HIV in clones J-Lat E27 and A2. Genome organization of UBXD8 and UTX genes and of the HIV integration in J-Lat E27 and A2 clones. (D) UBXD8 and UTX are active genes in Jurkat cells. RNA was extracted from growing Jurkat cells and used to measure expression of UBXD8, UTX, CXCR4, and PCAF genes by reverse transcription followed by real-time PCR of cDNA (RT-qPCR). In order to compare different amplicons, qPCR was performed in parallel from genomic DNA (gDNA). Data are expressed as relative units (RU) of cDNA amplification/gDNA amplification. Values represent the mean and standard deviation (SD) of a representative experiment performed in triplicate. (E) Effect of PMA treatment on UBXD8 and UTX genes. Jurkat or J-Lat cells were treated or not with PMA (10 nM) for 16 h, and RNA was extracted. UBXD8 and UTX expression was measured by RT-qPCR. GAPDH expression was measured for normalization. Data are expressed as fold change (FC) in expression in PMA-treated cells compared to that in untreated cells.