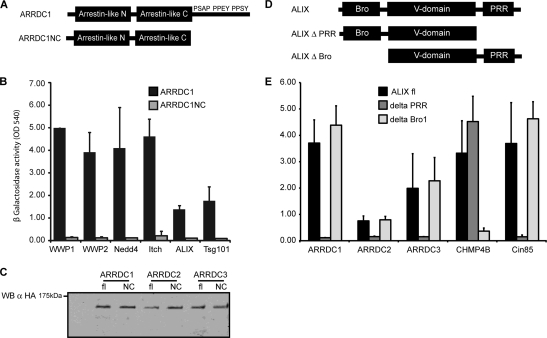
FIG. 3.
(A) Schematic drawing of ARRDC1 wild type (ARRDC1) and truncation mutant (ARRDC1NC) lacking the C terminus. (B) Binding of ARRDC1 to HECT ubiquitin ligases, ALIX, and Tsg101 is dependent on binding motifs in the C terminus. C-terminally truncated ARRDC1 fused to the Vp16 activation domain or GAL4 DNA binding domain was tested for its ability to interact with WWP1, WWP2, Nedd4, Itch, ALIX, and Tsg101 by yeast two-hybrid screening. Protein interactions were detected by measuring β-galactosidase activity in yeast lysates at an optical density at 540 nm (OD 540). Error bars indicate the standard deviations from the mean of two independent experiments measured in triplicates. (C) Expression of full-length (fl) and C-terminally truncated (NC) ARRDC1, ARRDC2, and ARRDC3 in yeast lysates. ARTs were expressed in the context of the HB18 vector (Vp16 activation domain), thereby generating an HA fusion protein. Protein expression was determined by lysis of yeast in protein sample buffer, followed by SDS-PAGE and Western blotting (WB) against HA. α, anti. (D) Schematic drawing of ALIX featuring the Bro1 and the V domain as well as the proline-rich region (PRR). (E) Binding of ARRDC1, ARRDC2, and ARRDC3 to ALIX is dependent on the proline-rich region in Alix. ARTs, CHMP4B, and Cin85 expressed as fusion proteins to the GAL4 DNA binding domain were tested for binding to ALIX by yeast two-hybrid screening as described for panel A.