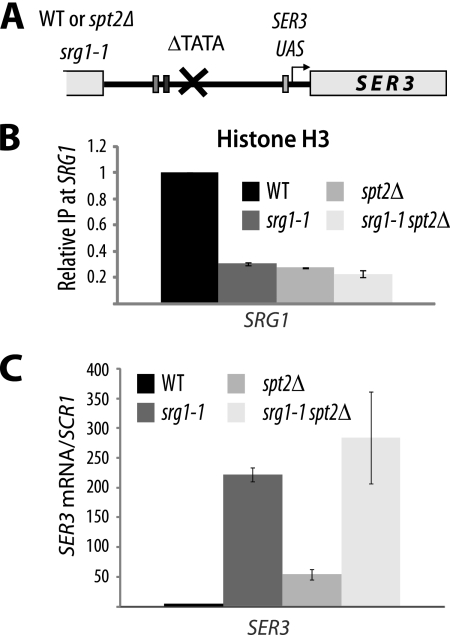
FIG. 8.
Chromatin structure at SRG1 depends on active transcription. (A) Diagram showing the srg1-1 construct. WT, wild type. (B) Mutation of SRG1 TATA is associated with a significant loss of histone H3 at the SRG1 3′-end region corresponding to the SER3 promoter. Histone H3 ChIP assays were conducted using chromatin extracted from wild-type (YAN1053), srg1-1 (YAN1054), spt2Δ (YAN1055), and srg1-1 spt2Δ (YAN1056) strains. For each strain, the value shown represents the ratio IP/input relative to the same ratio calculated for the corresponding wild-type strain and arbitrarily set at 1.0. The values shown are the averages and standard errors of results from three independent experiments. (C) The spt2Δ mutation in the srg1-1 strain does not increase significantly SER3 derepression. Total RNA extracted from wild-type (YAN1053), srg1-1 (YAN1054), spt2Δ (YAN1055), and srg1-1 spt2Δ (YAN1056) strains was used to produce cDNA that was subsequently quantified by qPCR. The SER3 mRNA relative level is the ratio of the SER3 mRNA to the SCR1 transcript level. The values shown are the averages and standard errors of results from three independent experiments.