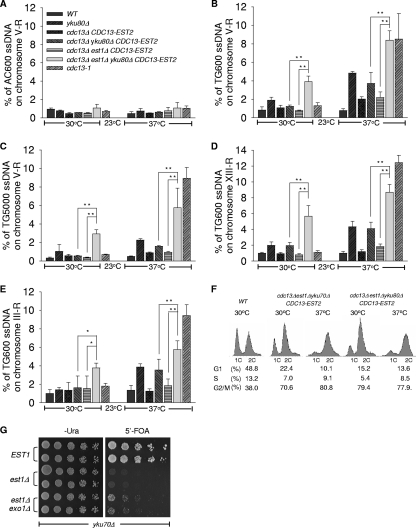
FIG. 2.
The est1Δ ykuΔ cells have increased single-stranded telomeric DNA. (A to E) QAOS analysis with yku80Δ mutants. Strains generated from 5′-FOA selection described in the legend to Fig. 1A and the wild-type strain were cultured at 30°C or 37°C for 4 h. cdc13-1 mutant strains cultured at 23 or 37°C were used as a positive control. Genomic DNA was extracted by the Zymolyase digestion method and monitored by QAOS analysis as described previously (3). The error bars represent the standard deviation (SD) from three independent experiments. **, P < 0.01; *, P < 0.05. (A) The relative amount of single-stranded CA DNA from the genomic locus 600 bp from the right-arm telomere in chromosome V (V-R) was determined. (B, D, and E) The relative amounts of single-stranded TG DNA from the genomic locus 600 bp from the right-arm telomeres in chromosome V (V-R) (B), chromosome XIII (XIII-R) (D), and chromosome III (III-R) (E) were determined. (C) The relative amount of single-stranded TG DNA from the genomic locus 5,000 bp from the telomere in the chromosome V right arm was determined. (F) FACS analysis with est1Δ ykuΔ mutant cells. Wild-type (WT) cells and the mutant strains indicated were cultured in selective liquid medium at 30 or 37°C for 4 h, and their DNA content was examined by FACS analysis. The percentages of cells in the G1, S, and G2/M phases are indicated at the bottom. (G) Growth analysis of est1Δ yku70Δ exo1Δ cells. est1Δ yku70Δ exo1Δ cells and est1Δ yku70Δ cells carrying plasmid pRS316-EST1 were plated at 30°C on medium selecting for the presence of pRS316-EST1 (Ura−) and on medium that selected for the eviction of covering EST1 plasmid (5′-FOA), respectively.