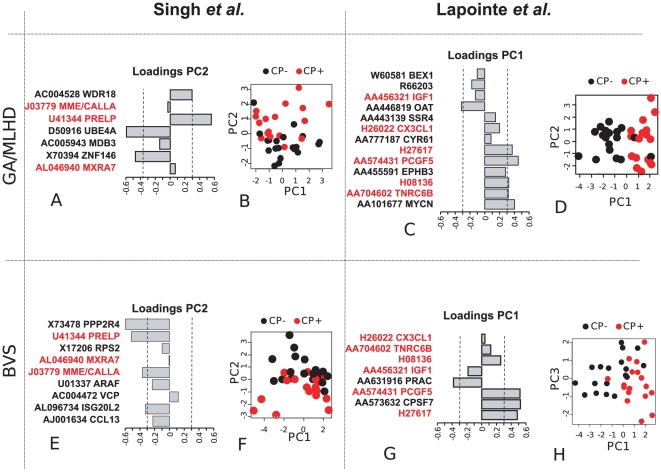
Figure 2. Principal component representation for Capsular Penetration using Normal Data.
The figure shows the result of a PCA representing sample separation on the basis of the expression in normal tissue of genes selected by the modelling procedures. Each quadrant in the figure represents a combination of a modelling approach and a specific dataset. Each quadrant contains a 2D plot representing the separation of capsular penetration negative (black close circles) and positive (red close circles) samples (plots B, D, F and H) and a bar chart (plots A, C, E and G) representing the PC loadings (x axis) for each gene component (y axis). Note that PC loadings represent the contribution of every gene to class separation. Dashed lines delimitated genes with larger contribution that are discussed in the manuscript. Genes present in GA-MLHD and BVS for the same dataset are highlighted in red.