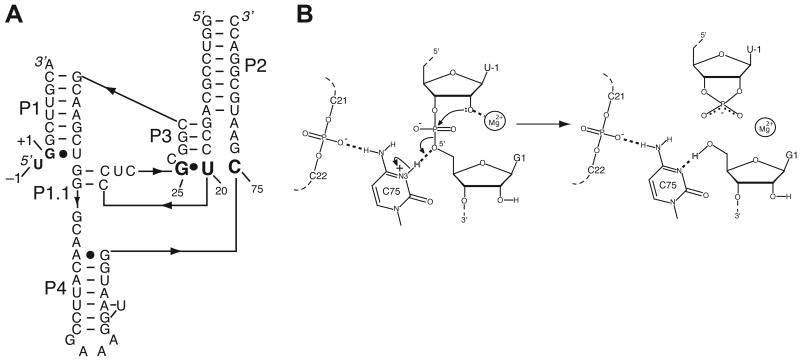
Figure 1.
Structure and proposed mechanism of the HDV ribozyme. (A) Secondary structure of the precleaved HDV ribozyme used in a recent crystallography study and herein for MD (PDB ID 3NKB). Numbering is based on the genomic HDV ribozyme (42). This is a two-piece, fast-folding U27Δ variant (90), in which P4 has been truncated and modified to facilitate crystallography (42). The reverse G•U wobble and catalytic C75 residue are in large bold font. The cleavage site is between U–1 and G1, and the five pairing regions are noted as P1–P4 and P1.1. The product ribozyme sequence used for MD is similar to the one shown, but it has a further truncated P4, a joining region between P1 and P2, a single additional nucleotide insertion, U27, and it lacks the –1 nucleotide (63). (B) Proposed mechanism of HDV ribozyme self-cleavage in which Mg2+ serves as a Lewis acid and C75 as a general acid (37, 39, 42). The C75 protonation states depicted in the precleaved and product states are used in the MD simulations.