Abstract
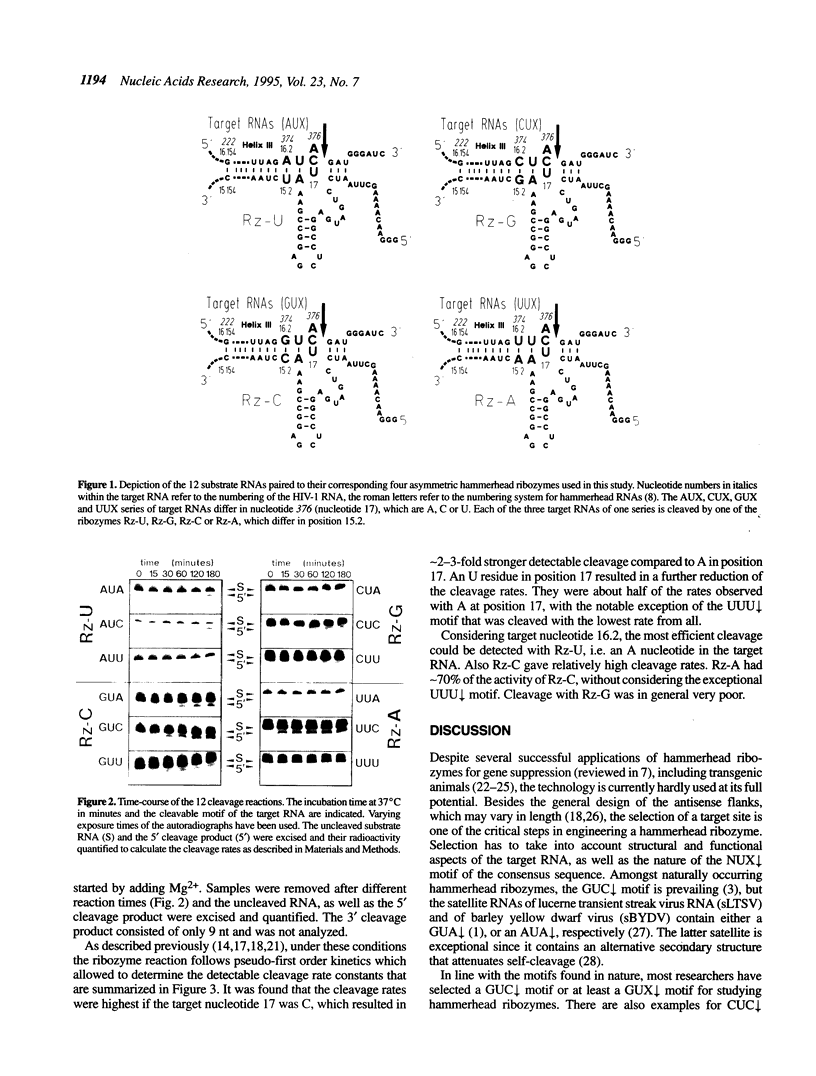
A trans-cleaving asymmetric hammerhead ribozyme directed against an AUC decreases target motif within an RNA specific for human immunodeficiency virus type 1 (HIV-1) was generated. The AUC decreases motif of the target RNA was permutated in order to generate all 12 variants of an NUX decreases consensus target motif, wherein N = A, C, G or U and X = A, C or U. Four asymmetric hammerhead ribozymes differing in the nucleotide that is complementary to N were generated, of which each was specific for three of the 12 target motifs. The residual sequence context within helices I and III remained unchanged. All 12 combinations resulted in cleavage of the target RNA. Using single-turnover conditions, the detectable cleavage rate constants at 37 degrees C were determined, which varied considerably depending on the NUX decreases motif. The NUC decreases motifs were cleaved more efficiently, with AUC decreases being cleaved best. Comparison with previous studies indicates that the sequence context of the NUX decreases motif plays a major role for the detectable cleavage activity.
Full text
PDF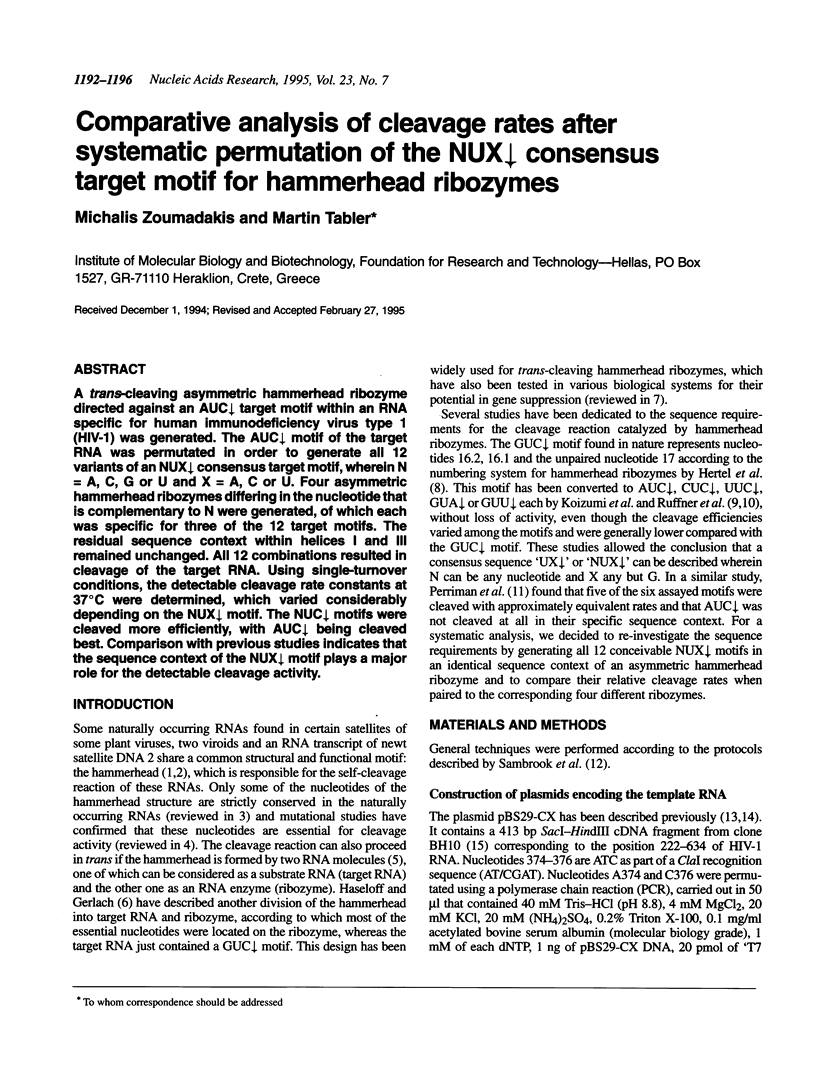



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bertrand E., Pictet R., Grange T. Can hammerhead ribozymes be efficient tools to inactivate gene function? Nucleic Acids Res. 1994 Feb 11;22(3):293–300. doi: 10.1093/nar/22.3.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruening G. Compilation of self-cleaving sequences from plant virus satellite RNAs and other sources. Methods Enzymol. 1989;180:546–558. doi: 10.1016/0076-6879(89)80123-5. [DOI] [PubMed] [Google Scholar]
- Cotten M., Schaffner G., Birnstiel M. L. Ribozyme, antisense RNA, and antisense DNA inhibition of U7 small nuclear ribonucleoprotein-mediated histone pre-mRNA processing in vitro. Mol Cell Biol. 1989 Oct;9(10):4479–4487. doi: 10.1128/mcb.9.10.4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisell P., Thompson S., James W. Inhibition of HIV-1 replication by ribozymes that show poor activity in vitro. Nucleic Acids Res. 1993 Nov 11;21(22):5251–5255. doi: 10.1093/nar/21.22.5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denman R. B. Cleavage of full-length beta APP mRNA by hammerhead ribozymes. Nucleic Acids Res. 1993 Aug 25;21(17):4119–4125. doi: 10.1093/nar/21.17.4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denman R. B. Using RNAFOLD to predict the activity of small catalytic RNAs. Biotechniques. 1993 Dec;15(6):1090–1095. [PubMed] [Google Scholar]
- Efrat S., Leiser M., Wu Y. J., Fusco-DeMane D., Emran O. A., Surana M., Jetton T. L., Magnuson M. A., Weir G., Fleischer N. Ribozyme-mediated attenuation of pancreatic beta-cell glucokinase expression in transgenic mice results in impaired glucose-induced insulin secretion. Proc Natl Acad Sci U S A. 1994 Mar 15;91(6):2051–2055. doi: 10.1073/pnas.91.6.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis J., Rogers J. Design and specificity of hammerhead ribozymes against calretinin mRNA. Nucleic Acids Res. 1993 Nov 11;21(22):5171–5178. doi: 10.1093/nar/21.22.5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein L. M., Pabón-Peña L. M. Alternative modes of self-cleavage by newt satellite 2 transcripts. Nucleic Acids Res. 1991 Apr 11;19(7):1699–1705. doi: 10.1093/nar/19.7.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster A. C., Symons R. H. Self-cleavage of plus and minus RNAs of a virusoid and a structural model for the active sites. Cell. 1987 Apr 24;49(2):211–220. doi: 10.1016/0092-8674(87)90562-9. [DOI] [PubMed] [Google Scholar]
- Forster A. C., Symons R. H. Self-cleavage of virusoid RNA is performed by the proposed 55-nucleotide active site. Cell. 1987 Jul 3;50(1):9–16. doi: 10.1016/0092-8674(87)90657-x. [DOI] [PubMed] [Google Scholar]
- Haseloff J., Gerlach W. L. Simple RNA enzymes with new and highly specific endoribonuclease activities. Nature. 1988 Aug 18;334(6183):585–591. doi: 10.1038/334585a0. [DOI] [PubMed] [Google Scholar]
- He Y. K., Lu C. D., Qi G. R. In vitro cleavage of HPV16 E6 and E7 RNA fragments by synthetic ribozymes and transcribed ribozymes from RNA-trimming plasmids. FEBS Lett. 1993 May 3;322(1):21–24. doi: 10.1016/0014-5793(93)81102-6. [DOI] [PubMed] [Google Scholar]
- Heidenreich O., Eckstein F. Hammerhead ribozyme-mediated cleavage of the long terminal repeat RNA of human immunodeficiency virus type 1. J Biol Chem. 1992 Jan 25;267(3):1904–1909. [PubMed] [Google Scholar]
- Heinrich J. C., Tabler M., Louis C. Attenuation of white gene expression in transgenic Drosophila melanogaster: possible role of a catalytic antisense RNA. Dev Genet. 1993;14(4):258–265. doi: 10.1002/dvg.1020140403. [DOI] [PubMed] [Google Scholar]
- Hertel K. J., Pardi A., Uhlenbeck O. C., Koizumi M., Ohtsuka E., Uesugi S., Cedergren R., Eckstein F., Gerlach W. L., Hodgson R. Numbering system for the hammerhead. Nucleic Acids Res. 1992 Jun 25;20(12):3252–3252. doi: 10.1093/nar/20.12.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homann M., Tabler M., Tzortzakaki S., Sczakiel G. Extension of helix II of an HIV-1-directed hammerhead ribozyme with long antisense flanks does not alter kinetic parameters in vitro but causes loss of the inhibitory potential in living cells. Nucleic Acids Res. 1994 Sep 25;22(19):3951–3957. doi: 10.1093/nar/22.19.3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homann M., Tzortzakaki S., Rittner K., Sczakiel G., Tabler M. Incorporation of the catalytic domain of a hammerhead ribozyme into antisense RNA enhances its inhibitory effect on the replication of human immunodeficiency virus type 1. Nucleic Acids Res. 1993 Jun 25;21(12):2809–2814. doi: 10.1093/nar/21.12.2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi M., Iwai S., Ohtsuka E. Cleavage of specific sites of RNA by designed ribozymes. FEBS Lett. 1988 Nov 7;239(2):285–288. doi: 10.1016/0014-5793(88)80935-9. [DOI] [PubMed] [Google Scholar]
- Koizumi M., Iwai S., Ohtsuka E. Construction of a series of several self-cleaving RNA duplexes using synthetic 21-mers. FEBS Lett. 1988 Feb 15;228(2):228–230. doi: 10.1016/0014-5793(88)80004-8. [DOI] [PubMed] [Google Scholar]
- Larsson S., Hotchkiss G., Andäng M., Nyholm T., Inzunza J., Jansson I., Ahrlund-Richter L. Reduced beta 2-microglobulin mRNA levels in transgenic mice expressing a designed hammerhead ribozyme. Nucleic Acids Res. 1994 Jun 25;22(12):2242–2248. doi: 10.1093/nar/22.12.2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschall P., Thomson J. B., Eckstein F. Inhibition of gene expression with ribozymes. Cell Mol Neurobiol. 1994 Oct;14(5):523–538. doi: 10.1007/BF02088835. [DOI] [PubMed] [Google Scholar]
- McCall M. J., Hendry P., Jennings P. A. Minimal sequence requirements for ribozyme activity. Proc Natl Acad Sci U S A. 1992 Jul 1;89(13):5710–5714. doi: 10.1073/pnas.89.13.5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller W. A., Hercus T., Waterhouse P. M., Gerlach W. L. A satellite RNA of barley yellow dwarf virus contains a novel hammerhead structure in the self-cleavage domain. Virology. 1991 Aug;183(2):711–720. doi: 10.1016/0042-6822(91)91000-7. [DOI] [PubMed] [Google Scholar]
- Miller W. A., Silver S. L. Alternative tertiary structure attenuates self-cleavage of the ribozyme in the satellite RNA of barley yellow dwarf virus. Nucleic Acids Res. 1991 Oct 11;19(19):5313–5320. doi: 10.1093/nar/19.19.5313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamaye K. L., Eckstein F. AUA-cleaving hammerhead ribozymes: attempted selection for improved cleavage. Biochemistry. 1994 Feb 8;33(5):1271–1277. doi: 10.1021/bi00171a030. [DOI] [PubMed] [Google Scholar]
- Pachuk C. J., Yoon K., Moelling K., Coney L. R. Selective cleavage of bcr-abl chimeric RNAs by a ribozyme targeted to non-contiguous sequences. Nucleic Acids Res. 1994 Feb 11;22(3):301–307. doi: 10.1093/nar/22.3.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perreault J. P., Wu T. F., Cousineau B., Ogilvie K. K., Cedergren R. Mixed deoxyribo- and ribo-oligonucleotides with catalytic activity. Nature. 1990 Apr 5;344(6266):565–567. doi: 10.1038/344565a0. [DOI] [PubMed] [Google Scholar]
- Perriman R., Delves A., Gerlach W. L. Extended target-site specificity for a hammerhead ribozyme. Gene. 1992 Apr 15;113(2):157–163. doi: 10.1016/0378-1119(92)90391-2. [DOI] [PubMed] [Google Scholar]
- Rittner K., Burmester C., Sczakiel G. In vitro selection of fast-hybridizing and effective antisense RNAs directed against the human immunodeficiency virus type 1. Nucleic Acids Res. 1993 Mar 25;21(6):1381–1387. doi: 10.1093/nar/21.6.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruffner D. E., Stormo G. D., Uhlenbeck O. C. Sequence requirements of the hammerhead RNA self-cleavage reaction. Biochemistry. 1990 Nov 27;29(47):10695–10702. doi: 10.1021/bi00499a018. [DOI] [PubMed] [Google Scholar]
- Saxena S. K., Ackerman E. J. Ribozymes correctly cleave a model substrate and endogenous RNA in vivo. J Biol Chem. 1990 Oct 5;265(28):17106–17109. [PubMed] [Google Scholar]
- Sczakiel G., Pawlita M., Kleinheinz A. Specific inhibition of human immunodeficiency virus type 1 replication by RNA transcribed in sense and antisense orientation from the 5'-leader/gag region. Biochem Biophys Res Commun. 1990 Jun 15;169(2):643–651. doi: 10.1016/0006-291x(90)90379-2. [DOI] [PubMed] [Google Scholar]
- Symons R. H. Small catalytic RNAs. Annu Rev Biochem. 1992;61:641–671. doi: 10.1146/annurev.bi.61.070192.003233. [DOI] [PubMed] [Google Scholar]
- Tabler M., Homann M., Tzortzakaki S., Sczakiel G. A three-nucleotide helix I is sufficient for full activity of a hammerhead ribozyme: advantages of an asymmetric design. Nucleic Acids Res. 1994 Sep 25;22(19):3958–3965. doi: 10.1093/nar/22.19.3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabler M., Tsagris M. Catalytic antisense RNAs produced by incorporating ribozyme cassettes into cDNA. Gene. 1991 Dec 15;108(2):175–183. doi: 10.1016/0378-1119(91)90432-b. [DOI] [PubMed] [Google Scholar]
- Tuschl T., Eckstein F. Hammerhead ribozymes: importance of stem-loop II for activity. Proc Natl Acad Sci U S A. 1993 Aug 1;90(15):6991–6994. doi: 10.1073/pnas.90.15.6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlenbeck O. C. A small catalytic oligoribonucleotide. Nature. 1987 Aug 13;328(6131):596–600. doi: 10.1038/328596a0. [DOI] [PubMed] [Google Scholar]
- Zhao J. J., Pick L. Generating loss-of-function phenotypes of the fushi tarazu gene with a targeted ribozyme in Drosophila. Nature. 1993 Sep 30;365(6445):448–451. doi: 10.1038/365448a0. [DOI] [PubMed] [Google Scholar]
- Zoumadakis M., Neubert W. J., Tabler M. The influence of imperfectly paired helices I and III on the catalytic activity of hammerhead ribozymes. Nucleic Acids Res. 1994 Dec 11;22(24):5271–5278. doi: 10.1093/nar/22.24.5271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989 Apr 7;244(4900):48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]
- von Weizsäcker F., Blum H. E., Wands J. R. Cleavage of hepatitis B virus RNA by three ribozymes transcribed from a single DNA template. Biochem Biophys Res Commun. 1992 Dec 15;189(2):743–748. doi: 10.1016/0006-291x(92)92264-x. [DOI] [PubMed] [Google Scholar]



