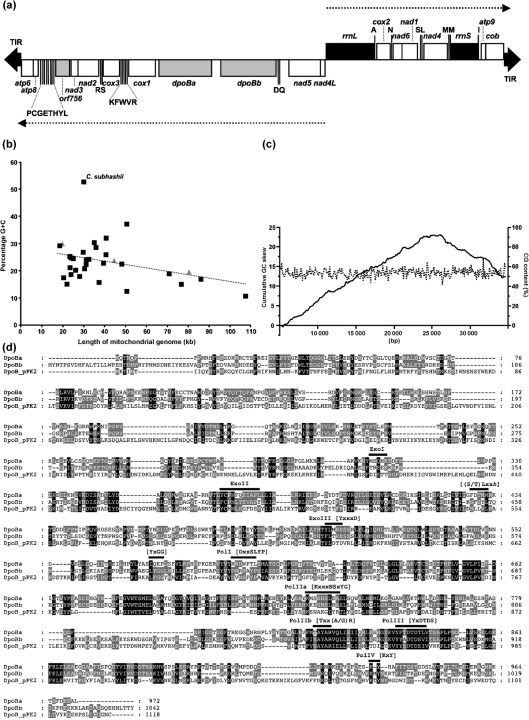
Fig. 1.
Linear mitochondrial genome from the yeast C. subhashii. (a) Genetic map of the 29 795 bp linear mtDNA. ORFs encoding proteins (open rectangles), rRNAs (black rectangles), tRNAs (labelled by the single-letter codes for their cognate amino acids) and long TIRs (black triangles at the termini) are indicated. Dotted lines with arrowheads indicate the direction of gene transcription. The genes presumably derived from a plasmid are shown as grey rectangles. Dubious ORFs mentioned in the text were omitted. (b) The percentage of G+C residues was plotted against the length of the mitochondrial genome. Available complete DNA sequences of yeast mitochondrial genomes were downloaded from public databases (Supplementary Table S1). Archiascomycete and hemiascomycete species are shown as grey triangles and black squares, respectively. (c) G+C content and cumulative GC skew analyses of the C. subhashii mtDNA were performed with window/step settings of 100/100. (d) Amino acid sequence alignment of putative DNA polymerases encoded by the C. subhashii mtDNA (DpoBa and DpoBb) and the linear plasmid pPK2 from mitochondria of Pichia kluyveri (DpoB_pPK2) was performed using the muscle utility (Edgar, 2004) of the Geneious Pro 4.8.5 package (Drummond et al., 2009) and manually adjusted. The shading was performed using the GeneDoc program (Nicholas et al., 1997). The motifs conserved in the family B DNA polymerases are shown above the sequences.