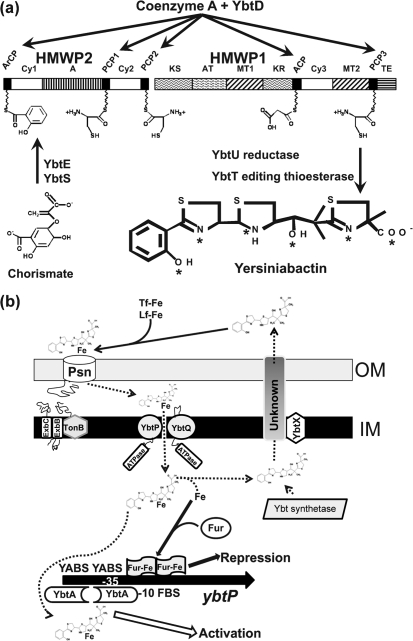
Fig. 1.
Models of yersiniabactin (Ybt) biosynthesis, transport and gene regulation. (a) Ybt biosynthesis. YbtS converts chorismate to salicylate and YbtE adenylates salicylate for attachment to HMWP2. YbtD transfers phosphopantetheinyl groups from coenzyme A to the indicated sites on HMWP2 and HMWP1 for attachment of salicylate, three cysteines and malonate. YbtU reduces one thiazolidine ring to a thiazoline ring, while YbtT is a TE that likely removes aberrant molecules from the Ybt synthetase enzyme complex. The TE domain of HMWP1 likely releases the completed siderophore from the enzyme complex. Asterisks in the Ybt structure indicate ferric ion coordination sites. HMWP2 and HMWP1 enzymatic domains: ArCP, aryl carrier protein; Cy, condensation/cyclization; A, adenylation; PCP, peptidyl carrier protein; KS, ketoacyl synthase; AT, acyltransferase; MT, methyltransferase; KR, β-ketoreductase; ACP, acyl carrier protein; TE, thioesterase. The model in (a) is reproduced with modifications from Perry & Fetherston (2004) with the permission of Horizon Scientific Press/Caister Academic Press, UK. (b) Model of Ybt transport and transcriptional regulation of ybtP. Synthesized Ybt is exported via an unknown mechanism that may include YbtX. Secreted Ybt can remove ferric iron from transferrin (Tf-Fe) and lactoferrin (Lf-Fe). We propose that the Ybt–Fe complex is transported through the outer membrane via the TonB-dependent receptor Psn. Ybt–Fe is transported through the inner membrane by YbtP/YbtQ. It is unclear whether this step requires a periplasmic or a membrane-spanning protein (none is shown here). In the cytoplasm, iron is released from Ybt by an unknown mechanism and is used nutritionally and for regulation of gene expression. The ybtP promoter, which controls expression of the ybtPQXS operon, is activated by YbtA, probably complexed with Ybt, and repressed by Fur in the presence of excess iron. Promoters for the irp2-irp1-ybtUTE and psn operons are similarly regulated. In contrast, YbtA negatively regulates transcription from its own promoter (Perry, 2004; Perry & Fetherston, 2004). YABS, YbtA binding site; FBS, Fur binding site; promoter region indicated by −10 and −35. Dashed arrows indicate steps or transported substrates that have not been experimentally determined.