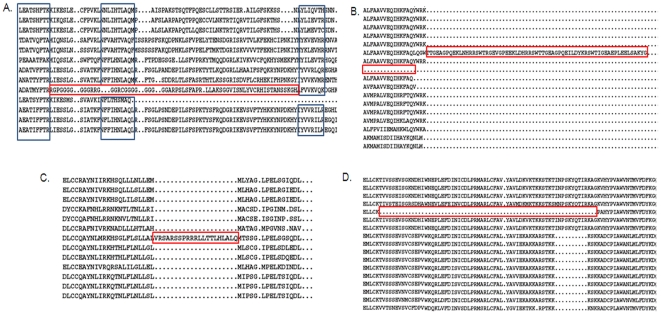
Figure 2. Examples of sequence discrepancies detected.
Four types of sequence discrepancies are identified and highlighted by red boxes in the subfamily alignments. A. Potential mispredicted exons are predicted based on the scores of the conserved core blocks (blue boxes) in the subfamily alignment. Here, the ninth sequence contains a segment ‘outlier’ that scores below the defined threshold for the central core block. The region of the sequence identified as a discrepancy is extended to the nearest core blocks in which the sequence is correctly aligned. B. Potential start and stop site errors are predicted based on the distribution of the positions of the N/C-terminal residues. C. Identification of a potential inserted intron, based on the presence of a single sequence with the insertion in a given subfamily. D. Identification of a potential missing exon, based on the presence of a single sequence with a deletion in a given subfamily.