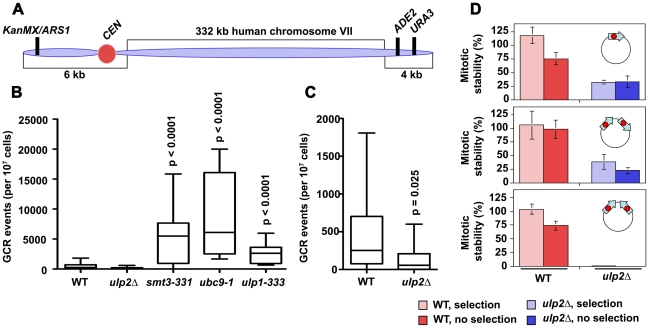
Figure 2. Chromosome re-arrangements in ulp2Δ mutants.
(A) The YAC used for GCR analysis consists of an origin of replication (ARS1), a CEN, and a long right arm containing 332 kbp of human DNA (adapted from [53]). GCRs deleting ADE2/URA3 at the terminus of right arm can be selected and distinguished from YAC loss through segregation errors. (B) WT (MLY068; 30 cultures), ulp2Δ (MLY069; 30 cultures), smt3-331 (MLY070; 10 cultures), ulp1-333 (MLY071; 10 cultures) and ubc9-1 (MLY072; 10 cultures) strains harboring the YAC were plated onto 5-FOA media to select for loss of URA3 and further genotyped to identify GCRs. Box plot graphs display median GCR events per 107 viable cells. p values (Student's t-test) were obtained from pair-wise comparisons between indicated mutants and the WT control. (C) Data as in B, but with lowered y-axis scale to show the reduction of GCRs in the ulp2Δ strain. It is possible to calculate that an average of 97.8±1.7% of WT cells and 93.6±4.4% of ulp2Δ cells retained the YAC at the time of plating to select for GCRs. (D) WT (CRY1) and ulp2Δ (JBY242) strains were transformed with three circular URA3 minichromosomes: monocentric p1XCEN, dicentric p2XCENdirect, and dicentric p2XCENinvert. Nine transformants for each strain/plasmid combination were cultured in parallel YPD (no selection for minichromosome) or Ura−/SC (to maintain selection) media, and equivalent volumes were plated onto YPD and Ura−/SC media. Graphs display the average percentage of cells retaining the minichromosome (mitotic stability), ± one standard deviation.